Gene
KWMTBOMO00544
Pre Gene Modal
BGIBMGA012335
Annotation
Transmembrane_protein_C2orf18-like_[Papilio_xuthus]
Full name
Solute carrier family 35 member F6
Location in the cell
PlasmaMembrane Reliability : 4.921
Sequence
CDS
ATGGCCTGGACAAACTACCAGAAGTTCCTCGCCCTGATGATGTTGGTGACTGGCTCAATCAACTCTTTGAGTACAAAATGGGCTGATATATTAAAATCAAAGGGGTCAGACGGTGAAGAAAGATATTTCAACCATCCATTCTTGCAGACCTTATCTATGTTTTTGGGTGAAATGCTATGTTTACTTGTGTTCAAATTGGTGTATTACAAAACACGGAACACTGGGGGACATTCATTAACAGCTGGAACACAAAACTTCAATCCCTTAATTCTGATGCCCGCAGCTATGTTGGATTTGATTGGAACATCCATAATGTATATTGGTTTGACTCTAACCTACATCAGCAGTTTCCAAATGTTCCGTGGGTCTATCATTGTCTTTGTGGCTGTGCTGTCCATGACATTCTCCAACAAACAGATCATCAAAAGAGAATGGCTCGGGATTGTGTGCGTCATCATCGGACTTTTGATCGTGGGAGTTACGGACGCTTTGTACCAAACACCGGGAGAGTCTAAAGGCCGCAACAGCCTTATAACCGGCGACCTACTGATTATAATCGCTCAGGTTGTATCCGCCTGTCAGATGGTGTACGAAGAGAGATACGTTACAGGTCAAAATATCCCTCCATTGCAAGCTGTAGGCTGGGAAGGGGTGTTCGGTTTCTCTGTGCTGTCCGGTCTGTTGGTCGTGTTCTACTGGACTCCGGCCCCGCCCCACTTCGACAACAACCCGAGGGGAACCATAGAAGACGCTATCGATGGCCTGATACAAATCGCCAACAATCCATTGATCCTCGTGGCAACGCTTGGTACGATTGTATCGATAGCTTTCTTCAATTTCGCCGGCATCAGCGTTACCAAGGAAATGTCTGCTACAACCAGAATGGTGCTCGACTCAGTGAGGTAA
Protein
MAWTNYQKFLALMMLVTGSINSLSTKWADILKSKGSDGEERYFNHPFLQTLSMFLGEMLCLLVFKLVYYKTRNTGGHSLTAGTQNFNPLILMPAAMLDLIGTSIMYIGLTLTYISSFQMFRGSIIVFVAVLSMTFSNKQIIKREWLGIVCVIIGLLIVGVTDALYQTPGESKGRNSLITGDLLIIIAQVVSACQMVYEERYVTGQNIPPLQAVGWEGVFGFSVLSGLLVVFYWTPAPPHFDNNPRGTIEDAIDGLIQIANNPLILVATLGTIVSIAFFNFAGISVTKEMSATTRMVLDSVR
Summary
Description
May play a role as a nucleotide-sugar transporter.
Subunit
Interacts with SLC25A5.
Similarity
Belongs to the SLC35F solute transporter family.
Feature
chain Solute carrier family 35 member F6
Uniprot
H9JS25
A0A2A4K439
A0A194QF62
A0A194QQD8
S4PT99
A0A212EKF5
+ More
A0A1B0DGQ6 A0A1B0CL98 A0A1L8DUU7 A0A023EPY1 A0A182GVT6 U5EUX3 Q17PC1 A0A1Q3FE67 D2A5T4 A0A1B6DHG4 B0WIW3 A0A1B6BZD9 A0A1B6KB83 A0A336M3N3 A0A0A9Y3L5 A0A1Y1LHV7 A0A1Z5KV46 A0A0P5KGZ1 A0A0P5KC74 A0A1J1HL08 A0A0P5FS89 A0A1D2NDJ6 A0A0N8AP30 Q7QAT4 A0A182IN62 A0A182S8S1 W5JX72 A0A2M4BRF9 L7M0A8 A0A2M3Z8F0 A0A023F9P5 A0A2M4AEG5 A0A0V0GCH5 A0A131YNK3 J3JUR2 A0A224XEJ2 V5I8K5 A0A224Z270 A0A1B6GK29 A0A1B6HVL9 A0A023FNA8 A0A2J7RA08 A0A293LZQ0 A0A023GP72 A0A131XCH7 A0A182MCM4 N6TRG2 A0A154PIS7 E9GF53 R4G451 A0A131XSN5 B7QC12 A0A0K8RJE4 A0A182Y7F8 A0A182KBX5 A0A182RZE3 A0A182F716 A0A2S2QH57 A0A2H8TMK8 J9JYA6 A0A1S3ITM5 A0A232ES29 F4W5V5 A0A2C9H8L8 A0A182UAV4 A0A1Y9GKQ6 A0A195EZM8 A0A158P2T8 K1R6G5 A0A2L2YA49 A0A182VS07 A0A151XHY1 A0A2A3EC14 A0A195C2A0 A0A026VXV3 A0A226EJI3 A0A147BA80 V5H2Q0 A0A3P8YBX1 A0A2S2PE48 A0A3B4C899 A0A087TXZ5 A0A060WCK3 A0A0P5IPC7 Q4RQY3 E2BZ82 H3DG54 A0A195BN35 G3UE46 H2V849 A0A060XMK4 A0A1S3M8Y8 A0A2Y9DNF8 H0W8X6
A0A1B0DGQ6 A0A1B0CL98 A0A1L8DUU7 A0A023EPY1 A0A182GVT6 U5EUX3 Q17PC1 A0A1Q3FE67 D2A5T4 A0A1B6DHG4 B0WIW3 A0A1B6BZD9 A0A1B6KB83 A0A336M3N3 A0A0A9Y3L5 A0A1Y1LHV7 A0A1Z5KV46 A0A0P5KGZ1 A0A0P5KC74 A0A1J1HL08 A0A0P5FS89 A0A1D2NDJ6 A0A0N8AP30 Q7QAT4 A0A182IN62 A0A182S8S1 W5JX72 A0A2M4BRF9 L7M0A8 A0A2M3Z8F0 A0A023F9P5 A0A2M4AEG5 A0A0V0GCH5 A0A131YNK3 J3JUR2 A0A224XEJ2 V5I8K5 A0A224Z270 A0A1B6GK29 A0A1B6HVL9 A0A023FNA8 A0A2J7RA08 A0A293LZQ0 A0A023GP72 A0A131XCH7 A0A182MCM4 N6TRG2 A0A154PIS7 E9GF53 R4G451 A0A131XSN5 B7QC12 A0A0K8RJE4 A0A182Y7F8 A0A182KBX5 A0A182RZE3 A0A182F716 A0A2S2QH57 A0A2H8TMK8 J9JYA6 A0A1S3ITM5 A0A232ES29 F4W5V5 A0A2C9H8L8 A0A182UAV4 A0A1Y9GKQ6 A0A195EZM8 A0A158P2T8 K1R6G5 A0A2L2YA49 A0A182VS07 A0A151XHY1 A0A2A3EC14 A0A195C2A0 A0A026VXV3 A0A226EJI3 A0A147BA80 V5H2Q0 A0A3P8YBX1 A0A2S2PE48 A0A3B4C899 A0A087TXZ5 A0A060WCK3 A0A0P5IPC7 Q4RQY3 E2BZ82 H3DG54 A0A195BN35 G3UE46 H2V849 A0A060XMK4 A0A1S3M8Y8 A0A2Y9DNF8 H0W8X6
Pubmed
19121390
26354079
23622113
22118469
24945155
26483478
+ More
17510324 18362917 19820115 25401762 26823975 28004739 28528879 27289101 12364791 14747013 17210077 20920257 23761445 25576852 25474469 26830274 22516182 28797301 28049606 23537049 21292972 25244985 28648823 21719571 21347285 22992520 26561354 24508170 30249741 25765539 25069045 24755649 15496914 20798317 21551351 21993624
17510324 18362917 19820115 25401762 26823975 28004739 28528879 27289101 12364791 14747013 17210077 20920257 23761445 25576852 25474469 26830274 22516182 28797301 28049606 23537049 21292972 25244985 28648823 21719571 21347285 22992520 26561354 24508170 30249741 25765539 25069045 24755649 15496914 20798317 21551351 21993624
EMBL
BABH01017106
BABH01017107
NWSH01000183
PCG78664.1
KQ459053
KPJ04183.1
+ More
KQ461181 KPJ07723.1 GAIX01012408 JAA80152.1 AGBW02014274 OWR41967.1 AJVK01060219 AJWK01017054 GFDF01003876 JAV10208.1 GAPW01001996 JAC11602.1 JXUM01002520 JXUM01002521 JXUM01002522 JXUM01002523 KQ560140 KXJ84216.1 GANO01002137 JAB57734.1 CH477193 EAT48510.1 GFDL01009184 JAV25861.1 KQ971345 EFA05027.1 GEDC01012175 JAS25123.1 DS231953 EDS28789.1 GEDC01030919 GEDC01028492 GEDC01021416 GEDC01021345 JAS06379.1 JAS08806.1 JAS15882.1 JAS15953.1 GEBQ01031277 GEBQ01027189 GEBQ01013425 GEBQ01004196 JAT08700.1 JAT12788.1 JAT26552.1 JAT35781.1 UFQT01000513 UFQT01000608 SSX24872.1 SSX25674.1 GBHO01015992 GDHC01011624 GDHC01003487 JAG27612.1 JAQ07005.1 JAQ15142.1 GEZM01057098 JAV72448.1 GFJQ02008013 JAV98956.1 GDIQ01209909 JAK41816.1 GDIP01172595 GDIQ01191668 LRGB01000248 JAJ50807.1 JAK60057.1 KZS20071.1 CVRI01000002 CRK86921.1 GDIQ01256820 JAJ94904.1 LJIJ01000077 ODN03330.1 GDIQ01256819 JAJ94905.1 AAAB01008888 EAA08806.3 ADMH02000081 ETN67874.1 GGFJ01006370 MBW55511.1 GACK01007273 JAA57761.1 GGFM01004058 MBW24809.1 GBBI01001069 JAC17643.1 GGFK01005853 MBW39174.1 GECL01000560 JAP05564.1 GEDV01008020 JAP80537.1 BT126977 AEE61939.1 GFTR01005539 JAW10887.1 GALX01004525 JAB63941.1 GFPF01012810 MAA23956.1 GECZ01031125 GECZ01006965 JAS38644.1 JAS62804.1 GECU01028959 GECU01027568 GECU01019078 GECU01017327 JAS78747.1 JAS80138.1 JAS88628.1 JAS90379.1 GBBK01001738 JAC22744.1 NEVH01006580 PNF37656.1 GFWV01008582 MAA33311.1 GBBM01000145 JAC35273.1 GEFH01003217 JAP65364.1 AXCM01001479 APGK01054667 KB741251 ENN71855.1 KQ434931 KZC11786.1 GL732541 EFX81958.1 ACPB03005214 GAHY01001650 JAA75860.1 GEFM01006581 JAP69215.1 ABJB010344060 ABJB010388403 DS903990 EEC16384.1 GADI01002598 JAA71210.1 GGMS01007865 MBY77068.1 GFXV01003415 MBW15220.1 ABLF02016845 NNAY01002506 OXU21170.1 GL887695 EGI70431.1 APCN01005504 KQ981905 KYN33367.1 ADTU01007478 JH816368 EKC41408.1 IAAA01012703 LAA05049.1 KQ982116 KYQ59921.1 KZ288308 PBC28739.1 KQ978344 KYM94989.1 KK107597 QOIP01000013 EZA48608.1 RLU15171.1 LNIX01000003 OXA56921.1 GEIB01000111 JAR87653.1 GANP01007203 JAB77265.1 GGMR01014577 MBY27196.1 KK117264 KFM69984.1 FR904422 CDQ62969.1 GDIQ01210564 JAK41161.1 CAAE01015003 CAG09199.1 GL451577 EFN78988.1 KQ976432 KYM87440.1 FR905292 CDQ78125.1 AAKN02023453
KQ461181 KPJ07723.1 GAIX01012408 JAA80152.1 AGBW02014274 OWR41967.1 AJVK01060219 AJWK01017054 GFDF01003876 JAV10208.1 GAPW01001996 JAC11602.1 JXUM01002520 JXUM01002521 JXUM01002522 JXUM01002523 KQ560140 KXJ84216.1 GANO01002137 JAB57734.1 CH477193 EAT48510.1 GFDL01009184 JAV25861.1 KQ971345 EFA05027.1 GEDC01012175 JAS25123.1 DS231953 EDS28789.1 GEDC01030919 GEDC01028492 GEDC01021416 GEDC01021345 JAS06379.1 JAS08806.1 JAS15882.1 JAS15953.1 GEBQ01031277 GEBQ01027189 GEBQ01013425 GEBQ01004196 JAT08700.1 JAT12788.1 JAT26552.1 JAT35781.1 UFQT01000513 UFQT01000608 SSX24872.1 SSX25674.1 GBHO01015992 GDHC01011624 GDHC01003487 JAG27612.1 JAQ07005.1 JAQ15142.1 GEZM01057098 JAV72448.1 GFJQ02008013 JAV98956.1 GDIQ01209909 JAK41816.1 GDIP01172595 GDIQ01191668 LRGB01000248 JAJ50807.1 JAK60057.1 KZS20071.1 CVRI01000002 CRK86921.1 GDIQ01256820 JAJ94904.1 LJIJ01000077 ODN03330.1 GDIQ01256819 JAJ94905.1 AAAB01008888 EAA08806.3 ADMH02000081 ETN67874.1 GGFJ01006370 MBW55511.1 GACK01007273 JAA57761.1 GGFM01004058 MBW24809.1 GBBI01001069 JAC17643.1 GGFK01005853 MBW39174.1 GECL01000560 JAP05564.1 GEDV01008020 JAP80537.1 BT126977 AEE61939.1 GFTR01005539 JAW10887.1 GALX01004525 JAB63941.1 GFPF01012810 MAA23956.1 GECZ01031125 GECZ01006965 JAS38644.1 JAS62804.1 GECU01028959 GECU01027568 GECU01019078 GECU01017327 JAS78747.1 JAS80138.1 JAS88628.1 JAS90379.1 GBBK01001738 JAC22744.1 NEVH01006580 PNF37656.1 GFWV01008582 MAA33311.1 GBBM01000145 JAC35273.1 GEFH01003217 JAP65364.1 AXCM01001479 APGK01054667 KB741251 ENN71855.1 KQ434931 KZC11786.1 GL732541 EFX81958.1 ACPB03005214 GAHY01001650 JAA75860.1 GEFM01006581 JAP69215.1 ABJB010344060 ABJB010388403 DS903990 EEC16384.1 GADI01002598 JAA71210.1 GGMS01007865 MBY77068.1 GFXV01003415 MBW15220.1 ABLF02016845 NNAY01002506 OXU21170.1 GL887695 EGI70431.1 APCN01005504 KQ981905 KYN33367.1 ADTU01007478 JH816368 EKC41408.1 IAAA01012703 LAA05049.1 KQ982116 KYQ59921.1 KZ288308 PBC28739.1 KQ978344 KYM94989.1 KK107597 QOIP01000013 EZA48608.1 RLU15171.1 LNIX01000003 OXA56921.1 GEIB01000111 JAR87653.1 GANP01007203 JAB77265.1 GGMR01014577 MBY27196.1 KK117264 KFM69984.1 FR904422 CDQ62969.1 GDIQ01210564 JAK41161.1 CAAE01015003 CAG09199.1 GL451577 EFN78988.1 KQ976432 KYM87440.1 FR905292 CDQ78125.1 AAKN02023453
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000092462
+ More
UP000092461 UP000069940 UP000249989 UP000008820 UP000007266 UP000002320 UP000076858 UP000183832 UP000094527 UP000007062 UP000075880 UP000075901 UP000000673 UP000235965 UP000075883 UP000019118 UP000076502 UP000000305 UP000015103 UP000001555 UP000076408 UP000075881 UP000075900 UP000069272 UP000007819 UP000085678 UP000215335 UP000007755 UP000076407 UP000075902 UP000075840 UP000078541 UP000005205 UP000005408 UP000075920 UP000075809 UP000242457 UP000078542 UP000053097 UP000279307 UP000198287 UP000265140 UP000261440 UP000054359 UP000193380 UP000008237 UP000007303 UP000078540 UP000007646 UP000005226 UP000087266 UP000248480 UP000005447
UP000092461 UP000069940 UP000249989 UP000008820 UP000007266 UP000002320 UP000076858 UP000183832 UP000094527 UP000007062 UP000075880 UP000075901 UP000000673 UP000235965 UP000075883 UP000019118 UP000076502 UP000000305 UP000015103 UP000001555 UP000076408 UP000075881 UP000075900 UP000069272 UP000007819 UP000085678 UP000215335 UP000007755 UP000076407 UP000075902 UP000075840 UP000078541 UP000005205 UP000005408 UP000075920 UP000075809 UP000242457 UP000078542 UP000053097 UP000279307 UP000198287 UP000265140 UP000261440 UP000054359 UP000193380 UP000008237 UP000007303 UP000078540 UP000007646 UP000005226 UP000087266 UP000248480 UP000005447
PRIDE
Pfam
Interpro
Gene 3D
ProteinModelPortal
H9JS25
A0A2A4K439
A0A194QF62
A0A194QQD8
S4PT99
A0A212EKF5
+ More
A0A1B0DGQ6 A0A1B0CL98 A0A1L8DUU7 A0A023EPY1 A0A182GVT6 U5EUX3 Q17PC1 A0A1Q3FE67 D2A5T4 A0A1B6DHG4 B0WIW3 A0A1B6BZD9 A0A1B6KB83 A0A336M3N3 A0A0A9Y3L5 A0A1Y1LHV7 A0A1Z5KV46 A0A0P5KGZ1 A0A0P5KC74 A0A1J1HL08 A0A0P5FS89 A0A1D2NDJ6 A0A0N8AP30 Q7QAT4 A0A182IN62 A0A182S8S1 W5JX72 A0A2M4BRF9 L7M0A8 A0A2M3Z8F0 A0A023F9P5 A0A2M4AEG5 A0A0V0GCH5 A0A131YNK3 J3JUR2 A0A224XEJ2 V5I8K5 A0A224Z270 A0A1B6GK29 A0A1B6HVL9 A0A023FNA8 A0A2J7RA08 A0A293LZQ0 A0A023GP72 A0A131XCH7 A0A182MCM4 N6TRG2 A0A154PIS7 E9GF53 R4G451 A0A131XSN5 B7QC12 A0A0K8RJE4 A0A182Y7F8 A0A182KBX5 A0A182RZE3 A0A182F716 A0A2S2QH57 A0A2H8TMK8 J9JYA6 A0A1S3ITM5 A0A232ES29 F4W5V5 A0A2C9H8L8 A0A182UAV4 A0A1Y9GKQ6 A0A195EZM8 A0A158P2T8 K1R6G5 A0A2L2YA49 A0A182VS07 A0A151XHY1 A0A2A3EC14 A0A195C2A0 A0A026VXV3 A0A226EJI3 A0A147BA80 V5H2Q0 A0A3P8YBX1 A0A2S2PE48 A0A3B4C899 A0A087TXZ5 A0A060WCK3 A0A0P5IPC7 Q4RQY3 E2BZ82 H3DG54 A0A195BN35 G3UE46 H2V849 A0A060XMK4 A0A1S3M8Y8 A0A2Y9DNF8 H0W8X6
A0A1B0DGQ6 A0A1B0CL98 A0A1L8DUU7 A0A023EPY1 A0A182GVT6 U5EUX3 Q17PC1 A0A1Q3FE67 D2A5T4 A0A1B6DHG4 B0WIW3 A0A1B6BZD9 A0A1B6KB83 A0A336M3N3 A0A0A9Y3L5 A0A1Y1LHV7 A0A1Z5KV46 A0A0P5KGZ1 A0A0P5KC74 A0A1J1HL08 A0A0P5FS89 A0A1D2NDJ6 A0A0N8AP30 Q7QAT4 A0A182IN62 A0A182S8S1 W5JX72 A0A2M4BRF9 L7M0A8 A0A2M3Z8F0 A0A023F9P5 A0A2M4AEG5 A0A0V0GCH5 A0A131YNK3 J3JUR2 A0A224XEJ2 V5I8K5 A0A224Z270 A0A1B6GK29 A0A1B6HVL9 A0A023FNA8 A0A2J7RA08 A0A293LZQ0 A0A023GP72 A0A131XCH7 A0A182MCM4 N6TRG2 A0A154PIS7 E9GF53 R4G451 A0A131XSN5 B7QC12 A0A0K8RJE4 A0A182Y7F8 A0A182KBX5 A0A182RZE3 A0A182F716 A0A2S2QH57 A0A2H8TMK8 J9JYA6 A0A1S3ITM5 A0A232ES29 F4W5V5 A0A2C9H8L8 A0A182UAV4 A0A1Y9GKQ6 A0A195EZM8 A0A158P2T8 K1R6G5 A0A2L2YA49 A0A182VS07 A0A151XHY1 A0A2A3EC14 A0A195C2A0 A0A026VXV3 A0A226EJI3 A0A147BA80 V5H2Q0 A0A3P8YBX1 A0A2S2PE48 A0A3B4C899 A0A087TXZ5 A0A060WCK3 A0A0P5IPC7 Q4RQY3 E2BZ82 H3DG54 A0A195BN35 G3UE46 H2V849 A0A060XMK4 A0A1S3M8Y8 A0A2Y9DNF8 H0W8X6
Ontologies
GO
PANTHER
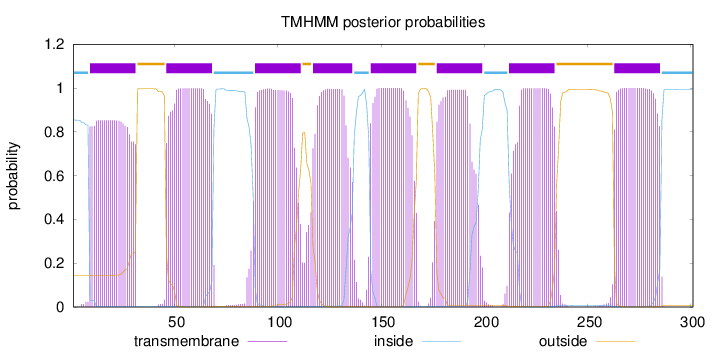
Topology
Subcellular location
Mitochondrion
Length:
301
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
169.46114
Exp number, first 60 AAs:
33.54596
Total prob of N-in:
0.85511
POSSIBLE N-term signal
sequence
inside
1 - 8
TMhelix
9 - 31
outside
32 - 45
TMhelix
46 - 68
inside
69 - 88
TMhelix
89 - 111
outside
112 - 116
TMhelix
117 - 136
inside
137 - 144
TMhelix
145 - 167
outside
168 - 176
TMhelix
177 - 199
inside
200 - 211
TMhelix
212 - 234
outside
235 - 262
TMhelix
263 - 285
inside
286 - 301
Population Genetic Test Statistics
Pi
16.030391
Theta
15.501496
Tajima's D
-1.359487
CLR
0.106565
CSRT
0.0778461076946153
Interpretation
Uncertain
