Gene
KWMTBOMO00543 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012334
Annotation
hypothetical_protein_KGM_21589_[Danaus_plexippus]
Location in the cell
Extracellular Reliability : 4.245
Sequence
CDS
ATGCGTATCCTCGGCATCTTAGCTGCTGTCGCCTGCTTTCAATTGGGCGTTTGGGCGTGTGAGCCCAATCAGGCTCAGAATGGGTGCAAAATATACGGTAACTCGTGCAGTTGTGGTTACGGTTGTAAAACTGAGTACATTTATAGGACAAGACGTTCTTGTTTGAATGCACTAAGAGAGAGAAGTTCAAACATATGCTCTCGTACGCCTTGCATTCGTGGCATATGCACGCAGACAATCCAGGATCCCGGATTCACTTGTAAATGCGAAGGCACCGGGTATTATGGAAAAAGATGTGAAAGAATCTGTCCAGCTACAGCAATACCAGGCACGTTCTTCCCACATGAGTGCATCGTTATCTAA
Protein
MRILGILAAVACFQLGVWACEPNQAQNGCKIYGNSCSCGYGCKTEYIYRTRRSCLNALRERSSNICSRTPCIRGICTQTIQDPGFTCKCEGTGYYGKRCERICPATAIPGTFFPHECIVI
Summary
Uniprot
H9JS24
A0A212EKJ0
A0A2A4K3Q7
I4DLF2
I4DLF1
A0A194QN56
+ More
A0A2Z5U754 A0A182UJ54 A0A182LDU6 A0A3F2Z210 A0A1S4GMM3 A0A182VLJ3 A0A182HQJ3 A0A182YKR2 A0NE78 A0A182RTV1 Q1HQE8 A0A182JSG4 A0A1W7R7B6 A0A1W7R7G4 A0A182PEN0 A0A182HDT0 A0A182WM00 A0A182FL99 U5ENH4 A0A2M3YWD0 T1EAH9 A0A151IHT5 A0A2M4DN41 W5J9J6 A0A2A3EJZ3 A0A195BCF1 A0A1B6DGW2 A0A182MLM5 A0A182IZ80 A0A0M9ADW8 A0A1B6CNP2 A0A084WCY6 A0A0C9RMS8 A0A1Q3FQY6 B0WET8 A0A1L8DBQ8 A0A1L8DP22 A0A1Y1MS17 A0A224XPK1 A0A171B3L3 D6WHZ0 F4WGX1 T1HDP3 A0A087ZZL1 A0A195FPL8 A0A151XFJ5 A0A195DAP3 A0A158NFF0 E2B5T3 A0A067R8T1 A0A2P8YW13 A0A026WHL6 A0A3L8DF69 A0A1J1IB92 E9G9G5 N6TRV3 A0A164NW85 E0W1Q0 E9IGH1 A0A232EXJ0 A0A0P5T8J8 A0A0P6BJI3 A0A0P4WQ20 A0A182N205 A0A0L7R994 E2ASR4 A0A1B6GP20 A0A1B6HGW9 A0A146L4Q3 A0A0A9ZEG0 A0A182QYN4 A0A2S2NPZ8 A0A336M8Z6 A0A2S2R9V4 C4WXJ6 A0A2H8TK30 K7IMD4 A0A0P4VHK9 T1JAV4 A0A2G8JNM1 R7V506 F6ZFF2 A0A0K2THV2 C3XWG9 A0A2C9LNC8 A0A2C9LJ32 A0A3M6TJW6 A0A2J8XT86 A0A2J8XT35 H2PEX9
A0A2Z5U754 A0A182UJ54 A0A182LDU6 A0A3F2Z210 A0A1S4GMM3 A0A182VLJ3 A0A182HQJ3 A0A182YKR2 A0NE78 A0A182RTV1 Q1HQE8 A0A182JSG4 A0A1W7R7B6 A0A1W7R7G4 A0A182PEN0 A0A182HDT0 A0A182WM00 A0A182FL99 U5ENH4 A0A2M3YWD0 T1EAH9 A0A151IHT5 A0A2M4DN41 W5J9J6 A0A2A3EJZ3 A0A195BCF1 A0A1B6DGW2 A0A182MLM5 A0A182IZ80 A0A0M9ADW8 A0A1B6CNP2 A0A084WCY6 A0A0C9RMS8 A0A1Q3FQY6 B0WET8 A0A1L8DBQ8 A0A1L8DP22 A0A1Y1MS17 A0A224XPK1 A0A171B3L3 D6WHZ0 F4WGX1 T1HDP3 A0A087ZZL1 A0A195FPL8 A0A151XFJ5 A0A195DAP3 A0A158NFF0 E2B5T3 A0A067R8T1 A0A2P8YW13 A0A026WHL6 A0A3L8DF69 A0A1J1IB92 E9G9G5 N6TRV3 A0A164NW85 E0W1Q0 E9IGH1 A0A232EXJ0 A0A0P5T8J8 A0A0P6BJI3 A0A0P4WQ20 A0A182N205 A0A0L7R994 E2ASR4 A0A1B6GP20 A0A1B6HGW9 A0A146L4Q3 A0A0A9ZEG0 A0A182QYN4 A0A2S2NPZ8 A0A336M8Z6 A0A2S2R9V4 C4WXJ6 A0A2H8TK30 K7IMD4 A0A0P4VHK9 T1JAV4 A0A2G8JNM1 R7V506 F6ZFF2 A0A0K2THV2 C3XWG9 A0A2C9LNC8 A0A2C9LJ32 A0A3M6TJW6 A0A2J8XT86 A0A2J8XT35 H2PEX9
Pubmed
19121390
22118469
22651552
26354079
20966253
12364791
+ More
25244985 17204158 17510324 26483478 20920257 23761445 24438588 28004739 18362917 19820115 21719571 21347285 20798317 24845553 29403074 24508170 30249741 21292972 23537049 20566863 21282665 28648823 26823975 25401762 20075255 27129103 29023486 23254933 12481130 15114417 18563158 15562597 30382153
25244985 17204158 17510324 26483478 20920257 23761445 24438588 28004739 18362917 19820115 21719571 21347285 20798317 24845553 29403074 24508170 30249741 21292972 23537049 20566863 21282665 28648823 26823975 25401762 20075255 27129103 29023486 23254933 12481130 15114417 18563158 15562597 30382153
EMBL
BABH01017105
AGBW02014274
OWR41968.1
NWSH01000183
PCG78666.1
AK402120
+ More
KQ459605 BAM18742.1 KPI91885.1 AK402119 BAM18741.1 KQ461196 KPJ06410.1 AP017491 BBB06758.1 AAAB01008948 APCN01004625 EAU76687.2 DQ440496 CH478476 ABF18529.1 EAT32911.1 GEHC01000625 JAV47020.1 GEHC01000620 JAV47025.1 JXUM01034790 KQ561036 KXJ79943.1 GANO01005044 JAB54827.1 GGFF01000103 MBW20570.1 GAMD01000392 JAB01199.1 KQ977578 KYN01816.1 GGFL01014794 MBW78972.1 ADMH02002077 ETN59530.1 KZ288230 PBC31582.1 KQ976514 KYM82228.1 GEDC01012367 JAS24931.1 AXCM01011595 AXCM01011596 KQ435691 KOX81149.1 GEDC01022208 JAS15090.1 ATLV01022899 KE525337 KFB48080.1 GBYB01009670 JAG79437.1 GFDL01005080 JAV29965.1 DS231911 EDS45851.1 GFDF01010314 JAV03770.1 GFDF01005933 JAV08151.1 GEZM01027275 JAV86786.1 GFTR01002010 JAW14416.1 GEMB01000314 JAS02805.1 KQ971331 EFA01024.1 GL888147 EGI66590.1 ACPB03029337 KQ981340 KYN42535.1 KQ982182 KYQ59153.1 KQ981063 KYN09922.1 ADTU01014382 GL445887 EFN88902.1 KK852630 KDR19870.1 PYGN01000327 PSN48442.1 KK107199 EZA55552.1 QOIP01000009 RLU18971.1 CVRI01000043 CRK96246.1 GL732536 EFX83879.1 APGK01057726 KB741280 KB632170 ENN71026.1 ERL89523.1 LRGB01002793 KZS06292.1 DS235873 EEB19632.1 GL763048 EFZ20304.1 NNAY01001740 OXU23063.1 GDIP01129510 JAL74204.1 GDIP01017074 JAM86641.1 GDRN01051525 JAI66391.1 KQ414627 KOC67404.1 GL442346 EFN63546.1 GECZ01005595 JAS64174.1 GECU01033847 JAS73859.1 GDHC01016492 JAQ02137.1 GBHO01001856 JAG41748.1 AXCN02000980 GGMR01006413 MBY19032.1 UFQS01000259 UFQT01000259 SSX02141.1 SSX22518.1 GGMS01017562 MBY86765.1 ABLF02033793 AK342675 BAH72616.1 GFXV01002594 MBW14399.1 GDKW01003413 JAI53182.1 JH432004 MRZV01001520 PIK37323.1 AMQN01005076 KB295063 ELU13634.1 EAAA01001480 HACA01008148 CDW25509.1 GG666471 EEN67703.1 RCHS01003480 RMX41534.1 NDHI03003320 PNJ85204.1 PNJ85205.1 ABGA01023905 ABGA01023906 ABGA01023907 ABGA01023908 ABGA01023909 ABGA01023910 ABGA01023911 ABGA01023912 ABGA01023913 ABGA01023914
KQ459605 BAM18742.1 KPI91885.1 AK402119 BAM18741.1 KQ461196 KPJ06410.1 AP017491 BBB06758.1 AAAB01008948 APCN01004625 EAU76687.2 DQ440496 CH478476 ABF18529.1 EAT32911.1 GEHC01000625 JAV47020.1 GEHC01000620 JAV47025.1 JXUM01034790 KQ561036 KXJ79943.1 GANO01005044 JAB54827.1 GGFF01000103 MBW20570.1 GAMD01000392 JAB01199.1 KQ977578 KYN01816.1 GGFL01014794 MBW78972.1 ADMH02002077 ETN59530.1 KZ288230 PBC31582.1 KQ976514 KYM82228.1 GEDC01012367 JAS24931.1 AXCM01011595 AXCM01011596 KQ435691 KOX81149.1 GEDC01022208 JAS15090.1 ATLV01022899 KE525337 KFB48080.1 GBYB01009670 JAG79437.1 GFDL01005080 JAV29965.1 DS231911 EDS45851.1 GFDF01010314 JAV03770.1 GFDF01005933 JAV08151.1 GEZM01027275 JAV86786.1 GFTR01002010 JAW14416.1 GEMB01000314 JAS02805.1 KQ971331 EFA01024.1 GL888147 EGI66590.1 ACPB03029337 KQ981340 KYN42535.1 KQ982182 KYQ59153.1 KQ981063 KYN09922.1 ADTU01014382 GL445887 EFN88902.1 KK852630 KDR19870.1 PYGN01000327 PSN48442.1 KK107199 EZA55552.1 QOIP01000009 RLU18971.1 CVRI01000043 CRK96246.1 GL732536 EFX83879.1 APGK01057726 KB741280 KB632170 ENN71026.1 ERL89523.1 LRGB01002793 KZS06292.1 DS235873 EEB19632.1 GL763048 EFZ20304.1 NNAY01001740 OXU23063.1 GDIP01129510 JAL74204.1 GDIP01017074 JAM86641.1 GDRN01051525 JAI66391.1 KQ414627 KOC67404.1 GL442346 EFN63546.1 GECZ01005595 JAS64174.1 GECU01033847 JAS73859.1 GDHC01016492 JAQ02137.1 GBHO01001856 JAG41748.1 AXCN02000980 GGMR01006413 MBY19032.1 UFQS01000259 UFQT01000259 SSX02141.1 SSX22518.1 GGMS01017562 MBY86765.1 ABLF02033793 AK342675 BAH72616.1 GFXV01002594 MBW14399.1 GDKW01003413 JAI53182.1 JH432004 MRZV01001520 PIK37323.1 AMQN01005076 KB295063 ELU13634.1 EAAA01001480 HACA01008148 CDW25509.1 GG666471 EEN67703.1 RCHS01003480 RMX41534.1 NDHI03003320 PNJ85204.1 PNJ85205.1 ABGA01023905 ABGA01023906 ABGA01023907 ABGA01023908 ABGA01023909 ABGA01023910 ABGA01023911 ABGA01023912 ABGA01023913 ABGA01023914
Proteomes
UP000005204
UP000007151
UP000218220
UP000053268
UP000053240
UP000075902
+ More
UP000075882 UP000076407 UP000075903 UP000075840 UP000076408 UP000007062 UP000075900 UP000008820 UP000075881 UP000075885 UP000069940 UP000249989 UP000075920 UP000069272 UP000078542 UP000000673 UP000242457 UP000078540 UP000075883 UP000075880 UP000053105 UP000030765 UP000002320 UP000007266 UP000007755 UP000015103 UP000005203 UP000078541 UP000075809 UP000078492 UP000005205 UP000008237 UP000027135 UP000245037 UP000053097 UP000279307 UP000183832 UP000000305 UP000019118 UP000030742 UP000076858 UP000009046 UP000215335 UP000075884 UP000053825 UP000000311 UP000075886 UP000007819 UP000002358 UP000230750 UP000014760 UP000008144 UP000001554 UP000076420 UP000275408 UP000001595
UP000075882 UP000076407 UP000075903 UP000075840 UP000076408 UP000007062 UP000075900 UP000008820 UP000075881 UP000075885 UP000069940 UP000249989 UP000075920 UP000069272 UP000078542 UP000000673 UP000242457 UP000078540 UP000075883 UP000075880 UP000053105 UP000030765 UP000002320 UP000007266 UP000007755 UP000015103 UP000005203 UP000078541 UP000075809 UP000078492 UP000005205 UP000008237 UP000027135 UP000245037 UP000053097 UP000279307 UP000183832 UP000000305 UP000019118 UP000030742 UP000076858 UP000009046 UP000215335 UP000075884 UP000053825 UP000000311 UP000075886 UP000007819 UP000002358 UP000230750 UP000014760 UP000008144 UP000001554 UP000076420 UP000275408 UP000001595
PRIDE
Pfam
Interpro
IPR000742
EGF-like_dom
+ More
IPR020683 Ankyrin_rpt-contain_dom
IPR038867 WAC
IPR036770 Ankyrin_rpt-contain_sf
IPR002110 Ankyrin_rpt
IPR026828 Suppressor_APCD_1/2
IPR006809 TAFII28
IPR009072 Histone-fold
IPR020846 MFS_dom
IPR011701 MFS
IPR036259 MFS_trans_sf
IPR013032 EGF-like_CS
IPR036249 Thioredoxin-like_sf
IPR031305 Casein_CS
IPR001791 Laminin_G
IPR002126 Cadherin-like_dom
IPR018097 EGF_Ca-bd_CS
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR020894 Cadherin_CS
IPR013320 ConA-like_dom_sf
IPR015919 Cadherin-like_sf
IPR001881 EGF-like_Ca-bd_dom
IPR009030 Growth_fac_rcpt_cys_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR038867 WAC
IPR036770 Ankyrin_rpt-contain_sf
IPR002110 Ankyrin_rpt
IPR026828 Suppressor_APCD_1/2
IPR006809 TAFII28
IPR009072 Histone-fold
IPR020846 MFS_dom
IPR011701 MFS
IPR036259 MFS_trans_sf
IPR013032 EGF-like_CS
IPR036249 Thioredoxin-like_sf
IPR031305 Casein_CS
IPR001791 Laminin_G
IPR002126 Cadherin-like_dom
IPR018097 EGF_Ca-bd_CS
IPR000152 EGF-type_Asp/Asn_hydroxyl_site
IPR020894 Cadherin_CS
IPR013320 ConA-like_dom_sf
IPR015919 Cadherin-like_sf
IPR001881 EGF-like_Ca-bd_dom
IPR009030 Growth_fac_rcpt_cys_sf
SUPFAM
Gene 3D
ProteinModelPortal
H9JS24
A0A212EKJ0
A0A2A4K3Q7
I4DLF2
I4DLF1
A0A194QN56
+ More
A0A2Z5U754 A0A182UJ54 A0A182LDU6 A0A3F2Z210 A0A1S4GMM3 A0A182VLJ3 A0A182HQJ3 A0A182YKR2 A0NE78 A0A182RTV1 Q1HQE8 A0A182JSG4 A0A1W7R7B6 A0A1W7R7G4 A0A182PEN0 A0A182HDT0 A0A182WM00 A0A182FL99 U5ENH4 A0A2M3YWD0 T1EAH9 A0A151IHT5 A0A2M4DN41 W5J9J6 A0A2A3EJZ3 A0A195BCF1 A0A1B6DGW2 A0A182MLM5 A0A182IZ80 A0A0M9ADW8 A0A1B6CNP2 A0A084WCY6 A0A0C9RMS8 A0A1Q3FQY6 B0WET8 A0A1L8DBQ8 A0A1L8DP22 A0A1Y1MS17 A0A224XPK1 A0A171B3L3 D6WHZ0 F4WGX1 T1HDP3 A0A087ZZL1 A0A195FPL8 A0A151XFJ5 A0A195DAP3 A0A158NFF0 E2B5T3 A0A067R8T1 A0A2P8YW13 A0A026WHL6 A0A3L8DF69 A0A1J1IB92 E9G9G5 N6TRV3 A0A164NW85 E0W1Q0 E9IGH1 A0A232EXJ0 A0A0P5T8J8 A0A0P6BJI3 A0A0P4WQ20 A0A182N205 A0A0L7R994 E2ASR4 A0A1B6GP20 A0A1B6HGW9 A0A146L4Q3 A0A0A9ZEG0 A0A182QYN4 A0A2S2NPZ8 A0A336M8Z6 A0A2S2R9V4 C4WXJ6 A0A2H8TK30 K7IMD4 A0A0P4VHK9 T1JAV4 A0A2G8JNM1 R7V506 F6ZFF2 A0A0K2THV2 C3XWG9 A0A2C9LNC8 A0A2C9LJ32 A0A3M6TJW6 A0A2J8XT86 A0A2J8XT35 H2PEX9
A0A2Z5U754 A0A182UJ54 A0A182LDU6 A0A3F2Z210 A0A1S4GMM3 A0A182VLJ3 A0A182HQJ3 A0A182YKR2 A0NE78 A0A182RTV1 Q1HQE8 A0A182JSG4 A0A1W7R7B6 A0A1W7R7G4 A0A182PEN0 A0A182HDT0 A0A182WM00 A0A182FL99 U5ENH4 A0A2M3YWD0 T1EAH9 A0A151IHT5 A0A2M4DN41 W5J9J6 A0A2A3EJZ3 A0A195BCF1 A0A1B6DGW2 A0A182MLM5 A0A182IZ80 A0A0M9ADW8 A0A1B6CNP2 A0A084WCY6 A0A0C9RMS8 A0A1Q3FQY6 B0WET8 A0A1L8DBQ8 A0A1L8DP22 A0A1Y1MS17 A0A224XPK1 A0A171B3L3 D6WHZ0 F4WGX1 T1HDP3 A0A087ZZL1 A0A195FPL8 A0A151XFJ5 A0A195DAP3 A0A158NFF0 E2B5T3 A0A067R8T1 A0A2P8YW13 A0A026WHL6 A0A3L8DF69 A0A1J1IB92 E9G9G5 N6TRV3 A0A164NW85 E0W1Q0 E9IGH1 A0A232EXJ0 A0A0P5T8J8 A0A0P6BJI3 A0A0P4WQ20 A0A182N205 A0A0L7R994 E2ASR4 A0A1B6GP20 A0A1B6HGW9 A0A146L4Q3 A0A0A9ZEG0 A0A182QYN4 A0A2S2NPZ8 A0A336M8Z6 A0A2S2R9V4 C4WXJ6 A0A2H8TK30 K7IMD4 A0A0P4VHK9 T1JAV4 A0A2G8JNM1 R7V506 F6ZFF2 A0A0K2THV2 C3XWG9 A0A2C9LNC8 A0A2C9LJ32 A0A3M6TJW6 A0A2J8XT86 A0A2J8XT35 H2PEX9
PDB
3POY
E-value=0.022358,
Score=81
Ontologies
GO
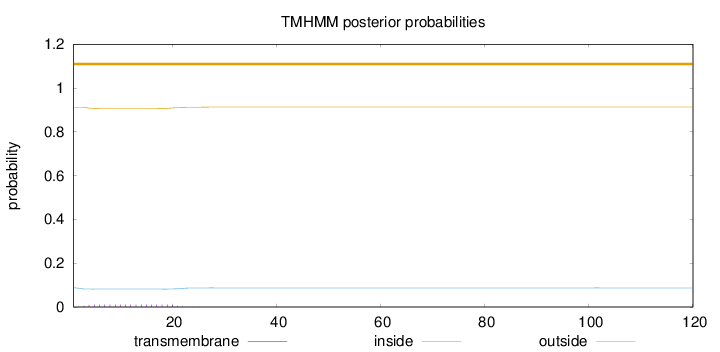
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.999669
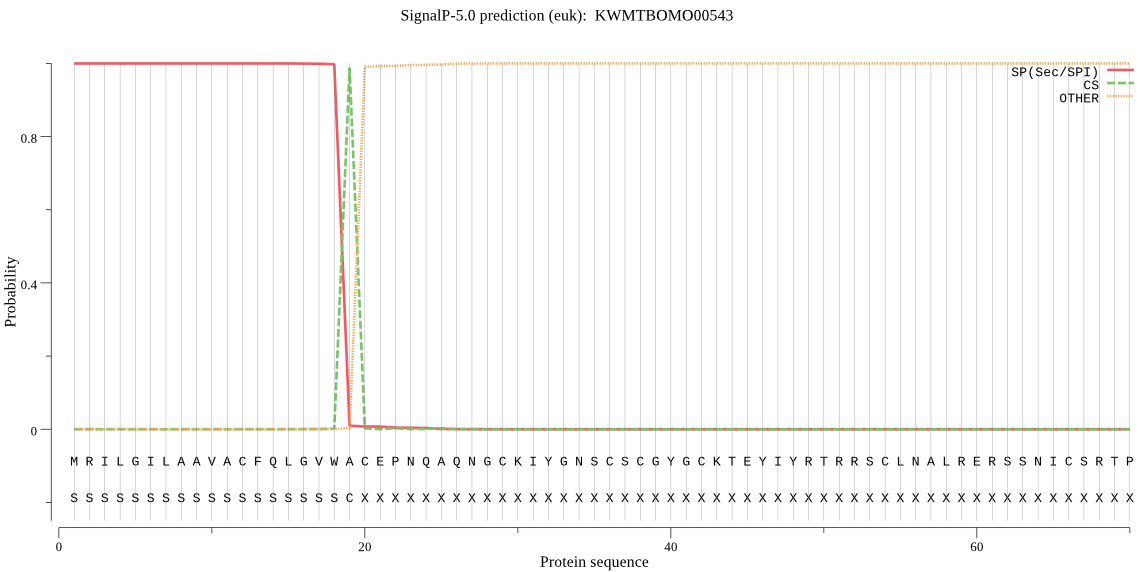
Length:
120
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.20877
Exp number, first 60 AAs:
0.20805
Total prob of N-in:
0.08777
outside
1 - 120
Population Genetic Test Statistics
Pi
14.653776
Theta
22.066654
Tajima's D
-0.732112
CLR
0.04102
CSRT
0.190940452977351
Interpretation
Uncertain
