Pre Gene Modal
BGIBMGA012333
Annotation
PREDICTED:_phytanoyl-CoA_dioxygenase_domain-containing_protein_1_[Bombyx_mori]
Location in the cell
Cytoplasmic Reliability : 1.625 Nuclear Reliability : 1.626
Sequence
CDS
ATGCACGACTCAATAAAGAGCCAGTTGATTACGGACGGCTACGCGATCCTAGAAGATTTTCTTCATGTTGCGGAATGTGATGAAATCAAAGCCGCTGGATTGGAATTCACTGAAAACTTGCCAGACATAGAGGAGAGAGCAACGTTCTCGACAACAGAGAAAACACATTTAAAGGATAAATATTTCCTAGAAAGCAACGACAAGATAAGGTGTTTCTTCGAAGAGGGAGCCATCGACGCCGATGGAAATCTTACAGTCGAACCGGAGATTTCATTAAACAAGGTCGGTCACGCACTGCATCTCCTACATCCGATTTTCAGATGTTACACGTATAGTGAACGCGTGAAGAGCATTTGCAAAGAGCTTGGCTTTATCGAGCCGGCTGTGGTGCAAAGCATGTATATTTTTAAGAACCCTGGCATTGGTTCCGAAGTGGTTCCACATCAAGACGCAACATATCTGTACACAGAGCCAACTCCCCCAGTTGGCTTCTGGATAGCTCTTGAAGAAGCCACAGTTCAAAACGGCTGTCTGTGGCTGTCTCGAGGCTCTCACAGGTCAGGAGTGCACAGACGTTTGATCCGCAACCCAGATGAAGACAGTGACGAGGCTTTAATCTATGATAAACCTGCAGCGGTTTATCCACAATCTAGCTTTACACCCATACCGGTTTCGAAAGGAACGTGTATATTATTACACGGCAACGTTGTACATAAGAGCTCCCACAACAAATCTGATAAGAGTCGCCACGCCTACACTTTCCACGTGGTGGAGACATACAATAATTTGTACTCGACCGAAAATTGGTTGCAGGAAGGCGAAGACGCTCCCTTTTGTAACTTGTACAACACATTACAGATGATTTAA
Protein
MHDSIKSQLITDGYAILEDFLHVAECDEIKAAGLEFTENLPDIEERATFSTTEKTHLKDKYFLESNDKIRCFFEEGAIDADGNLTVEPEISLNKVGHALHLLHPIFRCYTYSERVKSICKELGFIEPAVVQSMYIFKNPGIGSEVVPHQDATYLYTEPTPPVGFWIALEEATVQNGCLWLSRGSHRSGVHRRLIRNPDEDSDEALIYDKPAAVYPQSSFTPIPVSKGTCILLHGNVVHKSSHNKSDKSRHAYTFHVVETYNNLYSTENWLQEGEDAPFCNLYNTLQMI
Summary
Uniprot
H9JS23
A0A0B6VHE3
A0A2A4JKP4
I4DLF0
A0A194QS93
A0A194PGQ3
+ More
A0A0B6VIJ0 A0A212EKE9 A0A2H1V2R0 A0A2Z5ULY9 A0A2H1VG37 A0A2J7QTX7 A0A1B6GEC5 A0A067QJC2 A0A1B6IQ44 A0A023EMH8 Q17Q56 A0A1S4EUY1 Q17Q55 A0A182FL16 A0A088A5Q3 D6WGI8 A0A158NTQ7 A0A182HF21 W5J6Z6 A0A0J7L1L3 F4WJ01 Q7QAW1 A0A182VKF2 A0A182HZ45 A0A182WVZ4 A0A182KXA9 E2AIR3 A0A195FR23 B5DVF5 A0A0N0BD00 A0A195BP24 A0A182TIR7 A0A310S6Q9 A0A1L8DYY0 A0A182QXU4 A0A1Q3G4F6 A0A182NSJ7 A0A1B0BB94 A0A182VS42 A0A182P094 Q7QLE4 D3TPY0 A0A034WWF7 A0A0C9QDF8 A0A0K8UC17 A0A0L7QXZ9 A0A182M324 A0A195CV44 A0A084W1R7 A0A1A9Z8D7 A0A0L0CLL7 A0A232ETS2 A0A3B0K4P1 A0A182K2F6 A0A151X4Z5 A0A154PB32 A0A1A9UYY9 Q9VGY8 W8C8A5 B4LWF5 B4JIJ4 B4NF68 W8BQ75 B4PKQ9 B4K5K5 U5EY12 A0A1W4W0E2 B4HIS2 A0A1A9YFB5 A0A1B0CGX0 B3LVY0 B3P1M3 A0A1B0GEY0 A0A182JE58 B4G685 K7J4W9 A0A0C9PR10 A0A182YLL2 E2BBF5 A0A1L8DYU6 T1PI95 A0A026VYB4 A0A3L8D544 A0A182R898 A0A1J1J421 A0A2R7WS04 E0V9U6 B0X5I5 A0A1I8P4P4 A0A1Y1KXX9 J3JU81 T1HCC7 A0A224XLE9
A0A0B6VIJ0 A0A212EKE9 A0A2H1V2R0 A0A2Z5ULY9 A0A2H1VG37 A0A2J7QTX7 A0A1B6GEC5 A0A067QJC2 A0A1B6IQ44 A0A023EMH8 Q17Q56 A0A1S4EUY1 Q17Q55 A0A182FL16 A0A088A5Q3 D6WGI8 A0A158NTQ7 A0A182HF21 W5J6Z6 A0A0J7L1L3 F4WJ01 Q7QAW1 A0A182VKF2 A0A182HZ45 A0A182WVZ4 A0A182KXA9 E2AIR3 A0A195FR23 B5DVF5 A0A0N0BD00 A0A195BP24 A0A182TIR7 A0A310S6Q9 A0A1L8DYY0 A0A182QXU4 A0A1Q3G4F6 A0A182NSJ7 A0A1B0BB94 A0A182VS42 A0A182P094 Q7QLE4 D3TPY0 A0A034WWF7 A0A0C9QDF8 A0A0K8UC17 A0A0L7QXZ9 A0A182M324 A0A195CV44 A0A084W1R7 A0A1A9Z8D7 A0A0L0CLL7 A0A232ETS2 A0A3B0K4P1 A0A182K2F6 A0A151X4Z5 A0A154PB32 A0A1A9UYY9 Q9VGY8 W8C8A5 B4LWF5 B4JIJ4 B4NF68 W8BQ75 B4PKQ9 B4K5K5 U5EY12 A0A1W4W0E2 B4HIS2 A0A1A9YFB5 A0A1B0CGX0 B3LVY0 B3P1M3 A0A1B0GEY0 A0A182JE58 B4G685 K7J4W9 A0A0C9PR10 A0A182YLL2 E2BBF5 A0A1L8DYU6 T1PI95 A0A026VYB4 A0A3L8D544 A0A182R898 A0A1J1J421 A0A2R7WS04 E0V9U6 B0X5I5 A0A1I8P4P4 A0A1Y1KXX9 J3JU81 T1HCC7 A0A224XLE9
Pubmed
19121390
22651552
26354079
22118469
24845553
24945155
+ More
17510324 18362917 19820115 21347285 26483478 20920257 23761445 21719571 12364791 14747013 17210077 20966253 20798317 15632085 17994087 9087549 20353571 25348373 24438588 26108605 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24495485 17550304 20075255 25244985 25315136 24508170 30249741 20566863 28004739 22516182
17510324 18362917 19820115 21347285 26483478 20920257 23761445 21719571 12364791 14747013 17210077 20966253 20798317 15632085 17994087 9087549 20353571 25348373 24438588 26108605 28648823 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 24495485 17550304 20075255 25244985 25315136 24508170 30249741 20566863 28004739 22516182
EMBL
BABH01017102
AB926418
AB926420
BAQ19531.1
BAQ19533.1
NWSH01001117
+ More
PCG72531.1 AK402118 BAM18740.1 KQ461196 KPJ06411.1 KQ459605 KPI91884.1 AB926421 BAQ19534.1 AGBW02014274 OWR41969.1 ODYU01000405 SOQ35127.1 AP017491 BBB06757.1 ODYU01002396 SOQ39830.1 NEVH01011192 PNF32040.1 GECZ01008980 JAS60789.1 KK853424 KDR07687.1 GECU01018660 JAS89046.1 GAPW01003071 JAC10527.1 CH477187 EAT48861.1 EAT48860.1 KQ971321 EFA00565.1 ADTU01026110 JXUM01133286 KQ567959 KXJ69214.1 ADMH02002130 ETN58554.1 LBMM01001244 KMQ96657.1 GL888181 EGI65771.1 AAAB01008888 EAA08759.5 APCN01005482 GL439869 EFN66675.1 KQ981382 KYN42359.1 CM000070 EDY67741.2 KQ435891 KOX69391.1 KQ976435 KYM87109.1 KQ767283 OAD53424.1 GFDF01002403 JAV11681.1 AXCN02000234 GFDL01000350 JAV34695.1 JXJN01011342 JXJN01011343 AAAB01007485 EAA02969.4 EZ423482 ADD19758.1 GAKP01000819 JAC58133.1 GBYB01012473 GBYB01013172 JAG82240.1 JAG82939.1 GDHF01028130 JAI24184.1 KQ414699 KOC63419.1 AXCM01000846 KQ977279 KYN04039.1 ATLV01019415 KE525269 KFB44161.1 JRES01000228 KNC33190.1 NNAY01002235 OXU21755.1 OUUW01000013 SPP87672.1 KQ982526 KYQ55451.1 KQ434846 KZC08400.1 AE014297 AAF54534.3 GAMC01003129 JAC03427.1 CH940650 EDW66592.1 CH916369 EDV93075.1 CH964251 EDW82935.1 GAMC01003130 JAC03426.1 CM000160 EDW96818.1 CH933806 EDW14042.1 GANO01002247 JAB57624.1 CH480815 EDW42719.1 AJWK01011660 CH902617 EDV41513.1 CH954181 EDV49622.1 CCAG010012614 CCAG010012615 CCAG010012616 CCAG010012617 CCAG010012618 CCAG010012619 CH479179 EDW23842.1 GBYB01003678 JAG73445.1 GL447038 EFN86992.1 GFDF01002478 JAV11606.1 KA647643 AFP62272.1 KK107591 EZA48630.1 QOIP01000013 RLU15261.1 CVRI01000070 CRL07151.1 KK855392 PTY22342.1 DS234996 EEB10152.1 DS232381 EDS40870.1 GEZM01070921 GEZM01070920 GEZM01070916 GEZM01070915 JAV66224.1 BT126794 AEE61756.1 ACPB03008741 GFTR01004512 JAW11914.1
PCG72531.1 AK402118 BAM18740.1 KQ461196 KPJ06411.1 KQ459605 KPI91884.1 AB926421 BAQ19534.1 AGBW02014274 OWR41969.1 ODYU01000405 SOQ35127.1 AP017491 BBB06757.1 ODYU01002396 SOQ39830.1 NEVH01011192 PNF32040.1 GECZ01008980 JAS60789.1 KK853424 KDR07687.1 GECU01018660 JAS89046.1 GAPW01003071 JAC10527.1 CH477187 EAT48861.1 EAT48860.1 KQ971321 EFA00565.1 ADTU01026110 JXUM01133286 KQ567959 KXJ69214.1 ADMH02002130 ETN58554.1 LBMM01001244 KMQ96657.1 GL888181 EGI65771.1 AAAB01008888 EAA08759.5 APCN01005482 GL439869 EFN66675.1 KQ981382 KYN42359.1 CM000070 EDY67741.2 KQ435891 KOX69391.1 KQ976435 KYM87109.1 KQ767283 OAD53424.1 GFDF01002403 JAV11681.1 AXCN02000234 GFDL01000350 JAV34695.1 JXJN01011342 JXJN01011343 AAAB01007485 EAA02969.4 EZ423482 ADD19758.1 GAKP01000819 JAC58133.1 GBYB01012473 GBYB01013172 JAG82240.1 JAG82939.1 GDHF01028130 JAI24184.1 KQ414699 KOC63419.1 AXCM01000846 KQ977279 KYN04039.1 ATLV01019415 KE525269 KFB44161.1 JRES01000228 KNC33190.1 NNAY01002235 OXU21755.1 OUUW01000013 SPP87672.1 KQ982526 KYQ55451.1 KQ434846 KZC08400.1 AE014297 AAF54534.3 GAMC01003129 JAC03427.1 CH940650 EDW66592.1 CH916369 EDV93075.1 CH964251 EDW82935.1 GAMC01003130 JAC03426.1 CM000160 EDW96818.1 CH933806 EDW14042.1 GANO01002247 JAB57624.1 CH480815 EDW42719.1 AJWK01011660 CH902617 EDV41513.1 CH954181 EDV49622.1 CCAG010012614 CCAG010012615 CCAG010012616 CCAG010012617 CCAG010012618 CCAG010012619 CH479179 EDW23842.1 GBYB01003678 JAG73445.1 GL447038 EFN86992.1 GFDF01002478 JAV11606.1 KA647643 AFP62272.1 KK107591 EZA48630.1 QOIP01000013 RLU15261.1 CVRI01000070 CRL07151.1 KK855392 PTY22342.1 DS234996 EEB10152.1 DS232381 EDS40870.1 GEZM01070921 GEZM01070920 GEZM01070916 GEZM01070915 JAV66224.1 BT126794 AEE61756.1 ACPB03008741 GFTR01004512 JAW11914.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000235965
+ More
UP000027135 UP000008820 UP000069272 UP000005203 UP000007266 UP000005205 UP000069940 UP000249989 UP000000673 UP000036403 UP000007755 UP000007062 UP000075903 UP000075840 UP000076407 UP000075882 UP000000311 UP000078541 UP000001819 UP000053105 UP000078540 UP000075902 UP000075886 UP000075884 UP000092460 UP000075920 UP000075885 UP000053825 UP000075883 UP000078542 UP000030765 UP000092445 UP000037069 UP000215335 UP000268350 UP000075881 UP000075809 UP000076502 UP000078200 UP000000803 UP000008792 UP000001070 UP000007798 UP000002282 UP000009192 UP000192221 UP000001292 UP000092443 UP000092461 UP000007801 UP000008711 UP000092444 UP000075880 UP000008744 UP000002358 UP000076408 UP000008237 UP000095301 UP000053097 UP000279307 UP000075900 UP000183832 UP000009046 UP000002320 UP000095300 UP000015103
UP000027135 UP000008820 UP000069272 UP000005203 UP000007266 UP000005205 UP000069940 UP000249989 UP000000673 UP000036403 UP000007755 UP000007062 UP000075903 UP000075840 UP000076407 UP000075882 UP000000311 UP000078541 UP000001819 UP000053105 UP000078540 UP000075902 UP000075886 UP000075884 UP000092460 UP000075920 UP000075885 UP000053825 UP000075883 UP000078542 UP000030765 UP000092445 UP000037069 UP000215335 UP000268350 UP000075881 UP000075809 UP000076502 UP000078200 UP000000803 UP000008792 UP000001070 UP000007798 UP000002282 UP000009192 UP000192221 UP000001292 UP000092443 UP000092461 UP000007801 UP000008711 UP000092444 UP000075880 UP000008744 UP000002358 UP000076408 UP000008237 UP000095301 UP000053097 UP000279307 UP000075900 UP000183832 UP000009046 UP000002320 UP000095300 UP000015103
Pfam
PF05721 PhyH
Interpro
IPR008775
Phytyl_CoA_dOase
ProteinModelPortal
H9JS23
A0A0B6VHE3
A0A2A4JKP4
I4DLF0
A0A194QS93
A0A194PGQ3
+ More
A0A0B6VIJ0 A0A212EKE9 A0A2H1V2R0 A0A2Z5ULY9 A0A2H1VG37 A0A2J7QTX7 A0A1B6GEC5 A0A067QJC2 A0A1B6IQ44 A0A023EMH8 Q17Q56 A0A1S4EUY1 Q17Q55 A0A182FL16 A0A088A5Q3 D6WGI8 A0A158NTQ7 A0A182HF21 W5J6Z6 A0A0J7L1L3 F4WJ01 Q7QAW1 A0A182VKF2 A0A182HZ45 A0A182WVZ4 A0A182KXA9 E2AIR3 A0A195FR23 B5DVF5 A0A0N0BD00 A0A195BP24 A0A182TIR7 A0A310S6Q9 A0A1L8DYY0 A0A182QXU4 A0A1Q3G4F6 A0A182NSJ7 A0A1B0BB94 A0A182VS42 A0A182P094 Q7QLE4 D3TPY0 A0A034WWF7 A0A0C9QDF8 A0A0K8UC17 A0A0L7QXZ9 A0A182M324 A0A195CV44 A0A084W1R7 A0A1A9Z8D7 A0A0L0CLL7 A0A232ETS2 A0A3B0K4P1 A0A182K2F6 A0A151X4Z5 A0A154PB32 A0A1A9UYY9 Q9VGY8 W8C8A5 B4LWF5 B4JIJ4 B4NF68 W8BQ75 B4PKQ9 B4K5K5 U5EY12 A0A1W4W0E2 B4HIS2 A0A1A9YFB5 A0A1B0CGX0 B3LVY0 B3P1M3 A0A1B0GEY0 A0A182JE58 B4G685 K7J4W9 A0A0C9PR10 A0A182YLL2 E2BBF5 A0A1L8DYU6 T1PI95 A0A026VYB4 A0A3L8D544 A0A182R898 A0A1J1J421 A0A2R7WS04 E0V9U6 B0X5I5 A0A1I8P4P4 A0A1Y1KXX9 J3JU81 T1HCC7 A0A224XLE9
A0A0B6VIJ0 A0A212EKE9 A0A2H1V2R0 A0A2Z5ULY9 A0A2H1VG37 A0A2J7QTX7 A0A1B6GEC5 A0A067QJC2 A0A1B6IQ44 A0A023EMH8 Q17Q56 A0A1S4EUY1 Q17Q55 A0A182FL16 A0A088A5Q3 D6WGI8 A0A158NTQ7 A0A182HF21 W5J6Z6 A0A0J7L1L3 F4WJ01 Q7QAW1 A0A182VKF2 A0A182HZ45 A0A182WVZ4 A0A182KXA9 E2AIR3 A0A195FR23 B5DVF5 A0A0N0BD00 A0A195BP24 A0A182TIR7 A0A310S6Q9 A0A1L8DYY0 A0A182QXU4 A0A1Q3G4F6 A0A182NSJ7 A0A1B0BB94 A0A182VS42 A0A182P094 Q7QLE4 D3TPY0 A0A034WWF7 A0A0C9QDF8 A0A0K8UC17 A0A0L7QXZ9 A0A182M324 A0A195CV44 A0A084W1R7 A0A1A9Z8D7 A0A0L0CLL7 A0A232ETS2 A0A3B0K4P1 A0A182K2F6 A0A151X4Z5 A0A154PB32 A0A1A9UYY9 Q9VGY8 W8C8A5 B4LWF5 B4JIJ4 B4NF68 W8BQ75 B4PKQ9 B4K5K5 U5EY12 A0A1W4W0E2 B4HIS2 A0A1A9YFB5 A0A1B0CGX0 B3LVY0 B3P1M3 A0A1B0GEY0 A0A182JE58 B4G685 K7J4W9 A0A0C9PR10 A0A182YLL2 E2BBF5 A0A1L8DYU6 T1PI95 A0A026VYB4 A0A3L8D544 A0A182R898 A0A1J1J421 A0A2R7WS04 E0V9U6 B0X5I5 A0A1I8P4P4 A0A1Y1KXX9 J3JU81 T1HCC7 A0A224XLE9
PDB
3OBZ
E-value=3.44277e-62,
Score=603
Ontologies
KEGG
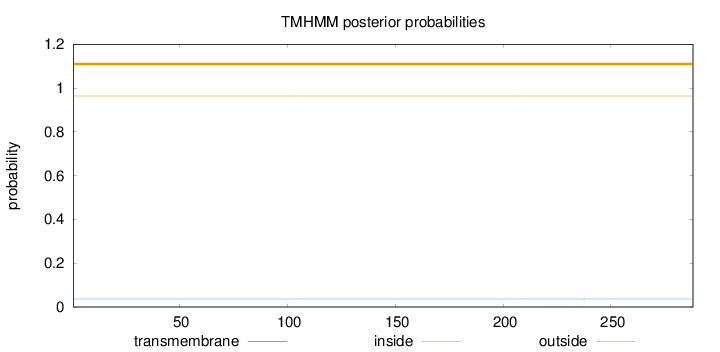
Topology
Length:
288
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00474
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03615
outside
1 - 288
Population Genetic Test Statistics
Pi
20.94102
Theta
16.198221
Tajima's D
-1.469178
CLR
0.023351
CSRT
0.0602969851507425
Interpretation
Uncertain
