Gene
KWMTBOMO00541
Pre Gene Modal
BGIBMGA012989
Annotation
Yokozuna_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.676
Sequence
CDS
ATGTTAGCCAGTCTGGATAAAGTTTTTGAGAGAAAAGGTATTTTCACTAAATTACACTTGCGAAGAAAGTTATTGTCTCTTAAGCTGAGGGATGATAAATTGGAAGACTATTTCATGAGATTTGAAAGTCTGATTAGAGAAATAGAAAATATTGATAAAAAGATGGATGAAGAAGTTAAAATATGTTATTTGCTGACGGGTATGTCGGAAAAGTACAACACAGTGATAACTGCTATTGAAACAATGTCTGCAGATAAAATTGACATGGAGTTTGTCAAGGCTCGGTTATTGGACGAAGAACTCAAACTGAATAATAAAGGTGAAGAGTATAATGATACAAAGAATGATGTAAGTTTTACTGCCTCAAGTGTAGAATGTTATCGTTGCCACAAAAAGGGCCATTTTGCACGTGAATGTAGAAGCAGAGGTCGTGGCTATTCAAGGAGAAACATTAATAATAAAGGTATATGTGGTAAAAACAGAACTGGAGAACAAAATAATATGACAGAAAGCATCCCAGAGAATAAATATAATTTTCTAGCAATAACGGAGTGTAATCTAAGTAGAGAAGACAAATTTATAGAATTTATAATAGATTCAGGAGCTACAGAAAATCTTATCAAGGAAGAATTAGAGAAATACATGTATGAAATTAAAAAACTACCAGATAAAATAAAAATTAAGATTGCAAATGGAGAAATATTAGAGGCTGACAAGAAAGGAAAGATAACCATAAGAGATAAAAATAATAATACTATAAATATAGAAGCGTTAATGGTCCCAGGGATTTATCATAATATAATATCTGTAAGAAACTTAAATATGAAAGGAAATAAAGTTGTTTTTACAAAATTTGGAGCACAAATAATAAATGGAAGAAAAGTCTTAAAATGTTCAGATAGAAATGGTTTATATATAGTACAAGGCGAAATAATACTAATAGAAGAAGCGAACTCAAGTGTACAGCAAGACAAAAATATTTGGCACCAACGTCTCGGACACTTAAATAGGAAATCATTAAAAATTATGGGTTTACCTGTGTCTGATGAAACATGTGGTCCTTGCATGAAGGGAAAATCTACTCGTTTATCATTCTATCCAGTTGACAGGCCAAGAAGTAGAAAAATAGGAGAATTATTGCATACAGATATTGGAGGTCCTGTATCACCAATTTCTTATGATGAAAAACAATATTACCAGACCATAACTGATGATTTTTCTCATTTTGTGACTGTTTATCTATTAAAGAATAAATCTGAAGCCACAAGAAATTTAATAAATCATATAAAGGAGTTAGAAAGACAAAAGGAAACTAAAGTTAAACGCATAAGATGTGATAATGGTGGAGAGTTCACCGCTGGCTGTTTGAAGCAATTTTGTATGGAAAATGGAATTATAATAGAGTATAGTCAACCTTACACACCACAACAAAATGGTGTGGCAGAGAGGCTAAACAGAACATTACTAGATAAGTCAAGAACTTTATTAAGTGAAACTGGATTACCTAAATATTTGTGGTCTGAGGCTATAATGTGTGCAGCATATCAATTGAATCGATGCCCATCAAGATCTATAAAGTATAAGATACCAGCTGAAATATACTACGGACTAGAAAATGTTAACTTGAACAAGTTAAAGGTATTTGGTTCTAAAGCATGGTATACGTGTGTCCCACAGAATAAGAAATTAGATAATAGAGCTGAAGAAGCCATTATGGTTGGCTATGGCAAACAAGGTTATAGATTATGGAATCCAAAAACAACAGAAATAATATTCTGCAGAGATGTCAAGTTTGATGAAAAGTACTATGAACATAAAAACAGTATTAAAAATAACAACTATATTTATTTACCAGAAGAGGACACACAAAATGAACTTATGTGCAATGATAAAAATAAAATTGAGGAAGAAAAAGTAATACAGAAAAATGAAGAGACAGACACAAAAAGAACTAGATCAGGAAGGATAATAAAGCAACCTGATTATCTAAAGAATTTTGGGACGTACACAGCATATTGTTACTTAGTGTGTAATGACCCAGAAAGCTATGAAGAAGCGATAAAAGAAGTTTCATGGAAAGAAGCGATACATAAGGAATTAGCAGCACTTGAGAACTTACAAAGCTGGGAAGAGACTAAACTACCGAAAGGAAAATTAGCTATAGACACTAAGTGGGTTTTTAGAACGAAAGACGATGGCACTAAGAAAGCCAGATTAGTCGCTCGAGGTTTTCAAATAAAAAAATTATGA
Protein
MLASLDKVFERKGIFTKLHLRRKLLSLKLRDDKLEDYFMRFESLIREIENIDKKMDEEVKICYLLTGMSEKYNTVITAIETMSADKIDMEFVKARLLDEELKLNNKGEEYNDTKNDVSFTASSVECYRCHKKGHFARECRSRGRGYSRRNINNKGICGKNRTGEQNNMTESIPENKYNFLAITECNLSREDKFIEFIIDSGATENLIKEELEKYMYEIKKLPDKIKIKIANGEILEADKKGKITIRDKNNNTINIEALMVPGIYHNIISVRNLNMKGNKVVFTKFGAQIINGRKVLKCSDRNGLYIVQGEIILIEEANSSVQQDKNIWHQRLGHLNRKSLKIMGLPVSDETCGPCMKGKSTRLSFYPVDRPRSRKIGELLHTDIGGPVSPISYDEKQYYQTITDDFSHFVTVYLLKNKSEATRNLINHIKELERQKETKVKRIRCDNGGEFTAGCLKQFCMENGIIIEYSQPYTPQQNGVAERLNRTLLDKSRTLLSETGLPKYLWSEAIMCAAYQLNRCPSRSIKYKIPAEIYYGLENVNLNKLKVFGSKAWYTCVPQNKKLDNRAEEAIMVGYGKQGYRLWNPKTTEIIFCRDVKFDEKYYEHKNSIKNNNYIYLPEEDTQNELMCNDKNKIEEEKVIQKNEETDTKRTRSGRIIKQPDYLKNFGTYTAYCYLVCNDPESYEEAIKEVSWKEAIHKELAALENLQSWEETKLPKGKLAIDTKWVFRTKDDGTKKARLVARGFQIKKL
Summary
Similarity
Belongs to the glutamine synthetase family.
Uniprot
O96968
A0A146L6N5
A0A0A9XG27
A0A0A9ZBW6
A0A0A9Z0I6
A0A146KTS1
+ More
A0A1B6DFK3 A0A0A9WLY1 A0A0A9WQA3 A0A0A9ZCB8 A0A146LV28 A0A182GHP7 A0A2S2PNK0 A0A1Y1L115 A0A182GA29 A0A0J7N374 A0A1Y1MP14 A0A1Y1M4K4 A0A1Y1MQF0 J9LXN4 A0A182GSW5 A0A1W7R6C3 J9KJ72 A0A0T6AT79 A0A182H4X3 A0A224XA26 A0A151RCJ0 A0A182G1G7 A0A023F102 A0A151TNK0 A0A151RCT9 A0A2N9EUG1 A0A2N9HSV3 A0A2N9GHB5 A0A2N9F1W0 A0A1W7R6K7 A0A1B6FK59 A0A0J7K6U3 A0A068B703 A0A2N9IT47 A0A2N9JB15 A0A2N9EE40 A0A2Z7CM54 A0A182GGE6 A0A2N9IKI1 A0A2N9J3Y8 A0A2N9IPG8 A0A1B6F1U1 A0A2N9GHK9 A0A2N9ESI4 A0A146LT61 A0A2N9GK98 A0A2N9HHD8 A0A2K3MYT4 A0A2A4JRI1 A0A2N9GTI4 A0A151IY32 A0A2N9JBD5 A0A0J7K689 A0A151TGQ2 A0A085MVP5 A0A0V0T3I0 A0A182GYI1 A5CBM1 A0A2G2VUS6 A0A151RBY1 A0A085N572 A0A0V0TUP0 A0A1Y1MJE9 A5AS37 E5SB91 A0A194RJV9 A0A151J9U8 A0A0A9XBM0 A0A151TPU3 A0A151RPT4 A5B5N4 A0A329RLJ1 A0A2K3N1R3 Q967L5 A0A151TGP0 A0A2Z6NJ75 A0A2Z6N0M9 A0A1B6C2D3 A0A2N9FNY4 A0A2N9IUL9 A0A2N9GXL8 B8YLY6 A0A2N9I1L5 A0A2A4J959 A0A2N9I4X9 A0A396INR0
A0A1B6DFK3 A0A0A9WLY1 A0A0A9WQA3 A0A0A9ZCB8 A0A146LV28 A0A182GHP7 A0A2S2PNK0 A0A1Y1L115 A0A182GA29 A0A0J7N374 A0A1Y1MP14 A0A1Y1M4K4 A0A1Y1MQF0 J9LXN4 A0A182GSW5 A0A1W7R6C3 J9KJ72 A0A0T6AT79 A0A182H4X3 A0A224XA26 A0A151RCJ0 A0A182G1G7 A0A023F102 A0A151TNK0 A0A151RCT9 A0A2N9EUG1 A0A2N9HSV3 A0A2N9GHB5 A0A2N9F1W0 A0A1W7R6K7 A0A1B6FK59 A0A0J7K6U3 A0A068B703 A0A2N9IT47 A0A2N9JB15 A0A2N9EE40 A0A2Z7CM54 A0A182GGE6 A0A2N9IKI1 A0A2N9J3Y8 A0A2N9IPG8 A0A1B6F1U1 A0A2N9GHK9 A0A2N9ESI4 A0A146LT61 A0A2N9GK98 A0A2N9HHD8 A0A2K3MYT4 A0A2A4JRI1 A0A2N9GTI4 A0A151IY32 A0A2N9JBD5 A0A0J7K689 A0A151TGQ2 A0A085MVP5 A0A0V0T3I0 A0A182GYI1 A5CBM1 A0A2G2VUS6 A0A151RBY1 A0A085N572 A0A0V0TUP0 A0A1Y1MJE9 A5AS37 E5SB91 A0A194RJV9 A0A151J9U8 A0A0A9XBM0 A0A151TPU3 A0A151RPT4 A5B5N4 A0A329RLJ1 A0A2K3N1R3 Q967L5 A0A151TGP0 A0A2Z6NJ75 A0A2Z6N0M9 A0A1B6C2D3 A0A2N9FNY4 A0A2N9IUL9 A0A2N9GXL8 B8YLY6 A0A2N9I1L5 A0A2A4J959 A0A2N9I4X9 A0A396INR0
Pubmed
EMBL
AB014676
BAA74713.1
GDHC01015802
JAQ02827.1
GBHO01027564
JAG16040.1
+ More
GBHO01002761 JAG40843.1 GBHO01004832 JAG38772.1 GDHC01019310 JAP99318.1 GEDC01012841 GEDC01000503 JAS24457.1 JAS36795.1 GBHO01035173 JAG08431.1 GBHO01034936 JAG08668.1 GBHO01002616 JAG40988.1 GDHC01007058 JAQ11571.1 JXUM01244080 KQ636179 KXJ62430.1 GGMR01018348 MBY30967.1 GEZM01072005 JAV65635.1 JXUM01155348 KQ572178 KXJ68066.1 LBMM01010924 KMQ87150.1 GEZM01028916 JAV86195.1 GEZM01042286 JAV79968.1 JAV86196.1 ABLF02003343 ABLF02041793 JXUM01085595 KQ563513 KXJ73717.1 GEHC01000928 JAV46717.1 LJIG01022881 KRT78250.1 JXUM01025857 KQ560736 KXJ81076.1 GFTR01008662 JAW07764.1 KQ483843 KYP40338.1 JXUM01136904 KQ568499 KXJ68982.1 GBBI01004033 JAC14679.1 CM003606 KYP68607.1 KYP40337.1 OIVN01000332 SPC78433.1 OIVN01003986 SPD14739.1 OIVN01001906 SPC98838.1 OIVN01000486 SPC81000.1 GEHC01000895 JAV46750.1 GECZ01019212 JAS50557.1 LBMM01012492 KMQ86193.1 KF740825 AIC77183.1 OIVN01006217 SPD28032.1 OIVN01006466 SPD33659.1 OIVN01000260 SPC77257.1 KQ995285 KZV47435.1 JXUM01011189 KQ560325 KXJ83029.1 OIVN01002424 OIVN01005946 SPD03496.1 SPD24589.1 OIVN01006353 SPD31284.1 OIVN01006155 SPD26398.1 GECZ01025604 JAS44165.1 OIVN01001931 SPC99063.1 OIVN01000279 SPC77544.1 GDHC01007678 JAQ10951.1 OIVN01002029 SPC99965.1 OIVN01003431 SPD11153.1 ASHM01014005 PNX95963.1 NWSH01000816 PCG74073.1 OIVN01002361 SPD02992.1 KQ980795 KYN13016.1 OIVN01006472 SPD33779.1 LBMM01012772 KMQ86003.1 CM003608 KYP66219.1 KL367628 KFD61291.1 JYDJ01000808 KRX33413.1 JXUM01097527 KQ564351 KXJ72390.1 AM489192 CAN71109.1 MLFT02000010 PHT36714.1 KQ483853 KYP40164.1 KL367553 KFD64618.1 JYDJ01000136 KRX42725.1 GEZM01032204 JAV84770.1 AM433482 CAN69340.1 KQ460297 KPJ16236.1 KQ979396 KYN21742.1 GBHO01026200 JAG17404.1 KYP69041.1 KQ483623 KYP44533.1 AM447500 CAN74198.1 MJFZ01000929 RAW24142.1 ASHM01015075 PNX96991.1 AY009101 AAK12626.1 KYP66220.1 DF974005 GAU43961.1 DF973299 GAU24969.1 GEDC01029908 JAS07390.1 OIVN01001308 SPC92446.1 OIVN01006211 SPD27863.1 OIVN01002513 SPD04258.1 FJ544856 ACL97386.1 OIVN01004651 SPD18552.1 NWSH01002292 PCG68657.1 PCG68658.1 OIVN01004824 SPD19458.1 PSQE01000003 RHN67289.1
GBHO01002761 JAG40843.1 GBHO01004832 JAG38772.1 GDHC01019310 JAP99318.1 GEDC01012841 GEDC01000503 JAS24457.1 JAS36795.1 GBHO01035173 JAG08431.1 GBHO01034936 JAG08668.1 GBHO01002616 JAG40988.1 GDHC01007058 JAQ11571.1 JXUM01244080 KQ636179 KXJ62430.1 GGMR01018348 MBY30967.1 GEZM01072005 JAV65635.1 JXUM01155348 KQ572178 KXJ68066.1 LBMM01010924 KMQ87150.1 GEZM01028916 JAV86195.1 GEZM01042286 JAV79968.1 JAV86196.1 ABLF02003343 ABLF02041793 JXUM01085595 KQ563513 KXJ73717.1 GEHC01000928 JAV46717.1 LJIG01022881 KRT78250.1 JXUM01025857 KQ560736 KXJ81076.1 GFTR01008662 JAW07764.1 KQ483843 KYP40338.1 JXUM01136904 KQ568499 KXJ68982.1 GBBI01004033 JAC14679.1 CM003606 KYP68607.1 KYP40337.1 OIVN01000332 SPC78433.1 OIVN01003986 SPD14739.1 OIVN01001906 SPC98838.1 OIVN01000486 SPC81000.1 GEHC01000895 JAV46750.1 GECZ01019212 JAS50557.1 LBMM01012492 KMQ86193.1 KF740825 AIC77183.1 OIVN01006217 SPD28032.1 OIVN01006466 SPD33659.1 OIVN01000260 SPC77257.1 KQ995285 KZV47435.1 JXUM01011189 KQ560325 KXJ83029.1 OIVN01002424 OIVN01005946 SPD03496.1 SPD24589.1 OIVN01006353 SPD31284.1 OIVN01006155 SPD26398.1 GECZ01025604 JAS44165.1 OIVN01001931 SPC99063.1 OIVN01000279 SPC77544.1 GDHC01007678 JAQ10951.1 OIVN01002029 SPC99965.1 OIVN01003431 SPD11153.1 ASHM01014005 PNX95963.1 NWSH01000816 PCG74073.1 OIVN01002361 SPD02992.1 KQ980795 KYN13016.1 OIVN01006472 SPD33779.1 LBMM01012772 KMQ86003.1 CM003608 KYP66219.1 KL367628 KFD61291.1 JYDJ01000808 KRX33413.1 JXUM01097527 KQ564351 KXJ72390.1 AM489192 CAN71109.1 MLFT02000010 PHT36714.1 KQ483853 KYP40164.1 KL367553 KFD64618.1 JYDJ01000136 KRX42725.1 GEZM01032204 JAV84770.1 AM433482 CAN69340.1 KQ460297 KPJ16236.1 KQ979396 KYN21742.1 GBHO01026200 JAG17404.1 KYP69041.1 KQ483623 KYP44533.1 AM447500 CAN74198.1 MJFZ01000929 RAW24142.1 ASHM01015075 PNX96991.1 AY009101 AAK12626.1 KYP66220.1 DF974005 GAU43961.1 DF973299 GAU24969.1 GEDC01029908 JAS07390.1 OIVN01001308 SPC92446.1 OIVN01006211 SPD27863.1 OIVN01002513 SPD04258.1 FJ544856 ACL97386.1 OIVN01004651 SPD18552.1 NWSH01002292 PCG68657.1 PCG68658.1 OIVN01004824 SPD19458.1 PSQE01000003 RHN67289.1
Proteomes
PRIDE
Pfam
Interpro
IPR036397
RNaseH_sf
+ More
IPR039537 Retrotran_Ty1/copia-like
IPR001584 Integrase_cat-core
IPR013103 RVT_2
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR025724 GAG-pre-integrase_dom
IPR021109 Peptidase_aspartic_dom_sf
IPR025314 DUF4219
IPR041588 Integrase_H2C2
IPR006912 Harbinger_derived_prot
IPR000719 Prot_kinase_dom
IPR008266 Tyr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR014746 Gln_synth/guanido_kin_cat_dom
IPR008146 Gln_synth_cat_dom
IPR039537 Retrotran_Ty1/copia-like
IPR001584 Integrase_cat-core
IPR013103 RVT_2
IPR012337 RNaseH-like_sf
IPR001878 Znf_CCHC
IPR036875 Znf_CCHC_sf
IPR025724 GAG-pre-integrase_dom
IPR021109 Peptidase_aspartic_dom_sf
IPR025314 DUF4219
IPR041588 Integrase_H2C2
IPR006912 Harbinger_derived_prot
IPR000719 Prot_kinase_dom
IPR008266 Tyr_kinase_AS
IPR011009 Kinase-like_dom_sf
IPR014746 Gln_synth/guanido_kin_cat_dom
IPR008146 Gln_synth_cat_dom
SUPFAM
Gene 3D
ProteinModelPortal
O96968
A0A146L6N5
A0A0A9XG27
A0A0A9ZBW6
A0A0A9Z0I6
A0A146KTS1
+ More
A0A1B6DFK3 A0A0A9WLY1 A0A0A9WQA3 A0A0A9ZCB8 A0A146LV28 A0A182GHP7 A0A2S2PNK0 A0A1Y1L115 A0A182GA29 A0A0J7N374 A0A1Y1MP14 A0A1Y1M4K4 A0A1Y1MQF0 J9LXN4 A0A182GSW5 A0A1W7R6C3 J9KJ72 A0A0T6AT79 A0A182H4X3 A0A224XA26 A0A151RCJ0 A0A182G1G7 A0A023F102 A0A151TNK0 A0A151RCT9 A0A2N9EUG1 A0A2N9HSV3 A0A2N9GHB5 A0A2N9F1W0 A0A1W7R6K7 A0A1B6FK59 A0A0J7K6U3 A0A068B703 A0A2N9IT47 A0A2N9JB15 A0A2N9EE40 A0A2Z7CM54 A0A182GGE6 A0A2N9IKI1 A0A2N9J3Y8 A0A2N9IPG8 A0A1B6F1U1 A0A2N9GHK9 A0A2N9ESI4 A0A146LT61 A0A2N9GK98 A0A2N9HHD8 A0A2K3MYT4 A0A2A4JRI1 A0A2N9GTI4 A0A151IY32 A0A2N9JBD5 A0A0J7K689 A0A151TGQ2 A0A085MVP5 A0A0V0T3I0 A0A182GYI1 A5CBM1 A0A2G2VUS6 A0A151RBY1 A0A085N572 A0A0V0TUP0 A0A1Y1MJE9 A5AS37 E5SB91 A0A194RJV9 A0A151J9U8 A0A0A9XBM0 A0A151TPU3 A0A151RPT4 A5B5N4 A0A329RLJ1 A0A2K3N1R3 Q967L5 A0A151TGP0 A0A2Z6NJ75 A0A2Z6N0M9 A0A1B6C2D3 A0A2N9FNY4 A0A2N9IUL9 A0A2N9GXL8 B8YLY6 A0A2N9I1L5 A0A2A4J959 A0A2N9I4X9 A0A396INR0
A0A1B6DFK3 A0A0A9WLY1 A0A0A9WQA3 A0A0A9ZCB8 A0A146LV28 A0A182GHP7 A0A2S2PNK0 A0A1Y1L115 A0A182GA29 A0A0J7N374 A0A1Y1MP14 A0A1Y1M4K4 A0A1Y1MQF0 J9LXN4 A0A182GSW5 A0A1W7R6C3 J9KJ72 A0A0T6AT79 A0A182H4X3 A0A224XA26 A0A151RCJ0 A0A182G1G7 A0A023F102 A0A151TNK0 A0A151RCT9 A0A2N9EUG1 A0A2N9HSV3 A0A2N9GHB5 A0A2N9F1W0 A0A1W7R6K7 A0A1B6FK59 A0A0J7K6U3 A0A068B703 A0A2N9IT47 A0A2N9JB15 A0A2N9EE40 A0A2Z7CM54 A0A182GGE6 A0A2N9IKI1 A0A2N9J3Y8 A0A2N9IPG8 A0A1B6F1U1 A0A2N9GHK9 A0A2N9ESI4 A0A146LT61 A0A2N9GK98 A0A2N9HHD8 A0A2K3MYT4 A0A2A4JRI1 A0A2N9GTI4 A0A151IY32 A0A2N9JBD5 A0A0J7K689 A0A151TGQ2 A0A085MVP5 A0A0V0T3I0 A0A182GYI1 A5CBM1 A0A2G2VUS6 A0A151RBY1 A0A085N572 A0A0V0TUP0 A0A1Y1MJE9 A5AS37 E5SB91 A0A194RJV9 A0A151J9U8 A0A0A9XBM0 A0A151TPU3 A0A151RPT4 A5B5N4 A0A329RLJ1 A0A2K3N1R3 Q967L5 A0A151TGP0 A0A2Z6NJ75 A0A2Z6N0M9 A0A1B6C2D3 A0A2N9FNY4 A0A2N9IUL9 A0A2N9GXL8 B8YLY6 A0A2N9I1L5 A0A2A4J959 A0A2N9I4X9 A0A396INR0
PDB
5UOQ
E-value=1.40092e-07,
Score=136
Ontologies
GO
PANTHER
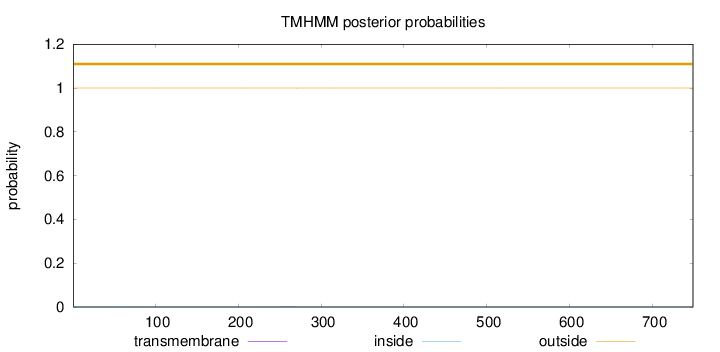
Topology
Length:
749
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00724999999999999
Exp number, first 60 AAs:
2e-05
Total prob of N-in:
0.00037
outside
1 - 749
Population Genetic Test Statistics
Pi
3.321286
Theta
4.764898
Tajima's D
-0.829215
CLR
0
CSRT
0.168891555422229
Interpretation
Uncertain
