Gene
KWMTBOMO00538
Pre Gene Modal
BGIBMGA012328
Annotation
PREDICTED:_proton-coupled_amino_acid_transporter_1_isoform_X1_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.923
Sequence
CDS
ATGATAGCCGAATATGATTCCAAGAAAAAAGGAGTCAGAAATGATTTATCCGATGTTGTTATGATCAAATACAATGTTGACCCAAGTGAGATCCCAGTGGAACAACAGGCGGGCTCGACCCTACCTCTTATGGAGATCCCGGGAAAGGATGTCGAGGCAGACGAGGACTATAATCCCTTCGAACACCGAAAACTTGCTCACCCTACCTCTGACATGGATACTTTAATTCACTTGCTGAAAGGTTCACTGGGCAGTGGTATTCTTGCCATGCCAATAGCATTCAAAAATGCTGGTCTTTTCTTCGGTCTGTTCGCGACTTTTGCCATCGGAGCTATCTGCACCTACTGCGTACATGTCCTGGTGAAAACTGCTCATGAGCTATGCAGACGCATGCAAAAGCCTTCATTAGGTTTCGCCGAAACAGCTGAAGCTGCTTTCTTATCTGGGCCGCCATCAATGCATAAATTTTCCAGACTCGCCAAAGCAATGGTTAACTGGTTCTTGGTATTAGATTTGCTGGGCTGTTGCTGCGTCTACATCGTATTCGTTTCCAAAAATGTAAGCCAAGTAGTTGACTACTACGCCGAAGGAACTTCATTCCACAATCTGCATATCCGAGTCTACATGGCTGCTCTTTTGCCATTCCTCATCATGATGAATCTTATTCGGAACTTGAAGTACTTAGCGCCTTTTTCTATGATTGCGAATGTATTGGTTGGAACTGGAATGGGAATCACCTTCTACTATCTATTCCAAGATATCCCTAGTGTGAGTGAAAGGGATTCCTTTGCTGAGATCTCAACCTTACCAATATTCTTCGGTACCGCTATCTTTGCCTTGGAGGGAATCGGCGTGGTGATGCCCTTGGAAAACAACATGAAGACTCCTACTCACTTCATTGGATGCCCCGGAGTATTGAACACTGGAATGTTTTTCGTCGTAACTCTCTACGCGTTCACAGGATTCTTTGGTTATCTGAAGTACGGAGATGAGACCAAAGGCAGTATTACCTTGAACTTACCGGAATCCGAGAAGCTGGGTCAATGTGTGAAACTTATGATTGCCACGGCCATATTCTTCACGTACAGTCTACAGTTCTACGTCCCAATGGAAATCATTTGGAACAACGTCCGTCACTGGTTTGGAGCTAAGAAGAATCTTGCCGAATACAGCATCAGGATCGCTTTGATTGTGATGACGGTTTGCATCGCTATAGCTATTCCGGATCTTGGACCATTCATTTCTTTAGTCGGGGCTGTCTGCTTATCATTCCTCGGATTGATCTTCCCTGCTATTATGGAAACTGTTACCTTCTGGGACCGACCGAACGGCTTGGGACGTTTCAATTGGGTTCTCTGGAAAAACGTATTCATGATCTCCTTCGGTATTCTCGGGTTCTTGACAGGTTCATATGTCAGCATCGTTGAAATTATTGAAGGATCTAGTAATTAA
Protein
MIAEYDSKKKGVRNDLSDVVMIKYNVDPSEIPVEQQAGSTLPLMEIPGKDVEADEDYNPFEHRKLAHPTSDMDTLIHLLKGSLGSGILAMPIAFKNAGLFFGLFATFAIGAICTYCVHVLVKTAHELCRRMQKPSLGFAETAEAAFLSGPPSMHKFSRLAKAMVNWFLVLDLLGCCCVYIVFVSKNVSQVVDYYAEGTSFHNLHIRVYMAALLPFLIMMNLIRNLKYLAPFSMIANVLVGTGMGITFYYLFQDIPSVSERDSFAEISTLPIFFGTAIFALEGIGVVMPLENNMKTPTHFIGCPGVLNTGMFFVVTLYAFTGFFGYLKYGDETKGSITLNLPESEKLGQCVKLMIATAIFFTYSLQFYVPMEIIWNNVRHWFGAKKNLAEYSIRIALIVMTVCIAIAIPDLGPFISLVGAVCLSFLGLIFPAIMETVTFWDRPNGLGRFNWVLWKNVFMISFGILGFLTGSYVSIVEIIEGSSN
Summary
Uniprot
H9JS18
S4P9N7
A0A0L7KRE4
A0A1E1WKV8
A0A212EKG6
A0A2H1VCT6
+ More
A0A194PFS3 A0A194QLI2 F4WE43 A0A151JRF8 A0A195BBC7 A0A154PBN8 A0A151JYP8 A0A151X3R2 A0A195CP10 E2BJJ1 A0A088AJT7 K7JB18 V9ID24 A0A2A3EI11 A0A0N0U7Q8 E2ASE4 A0A310S756 E9IG15 A0A0L7RK76 A0A0T6BEU7 A0A0C9RHK6 A0A1Y1NBW2 A0A1Y1NE78 A0A158NAG7 A0A1B6DQ99 A0A1B6E4V7 A0A1B6JE18 A0A1B6GFV2 V5I8J7 A0A1W4WXD7 A0A1B6KPD0 A0A1W4WYN7 A0A1B6KHZ3 A0A1L8DDW9 A0A1L8DDJ4 A0A2L0RID4 A0A2J7PW27 A0A1W4WMQ2 N6UDN4 A0A2M3YZF3 A0A084W8L1 U4U1N4 A0A2S2QYS6 A0A182KA57 A0A182KTL4 A0A182XJL1 A0A182VEC7 A0A1Q3FJZ8 A0A1Q3FJE8 A0A1Q3FJT2 A0A182TW42 A0A182HRI8 Q7PM43 A0A182YR05 A0A1Q3FKZ9 B0WVR2 A0A2C9GL31 A0A139WJW5 A0A1S4JWS2 A0A1B0CTW6 A0A182PCB0 A0A2L0RIE5 A0A0K8TQ57 A0A2H8TWY0 A0A182LX02 A0A182QCM2 A0A224XBK8 A0A0V0G3I9 A0A182RR74 A0A2M4CTL0 A0A2M4CTK4 W5JKP5 A0A2M4CTU7 A0A069DU01 A0A182NAR7 A0A182J8J2 A0A182W913 A0A2M4A4A2 A0A0A9YUE0 A0A3L8D5L8 T1EAB7 A0A182F3V1 A0A0P4VW95 T1E7Y8 A0A0A9YVX6 T1IBH5 A0A2M4A3X2 A0A0K8SD30 A0A182H342 A0A1Y0AWB0 A0A2M4BM04 Q173C1 A0A2M4BL63 A0A1S4FFU4
A0A194PFS3 A0A194QLI2 F4WE43 A0A151JRF8 A0A195BBC7 A0A154PBN8 A0A151JYP8 A0A151X3R2 A0A195CP10 E2BJJ1 A0A088AJT7 K7JB18 V9ID24 A0A2A3EI11 A0A0N0U7Q8 E2ASE4 A0A310S756 E9IG15 A0A0L7RK76 A0A0T6BEU7 A0A0C9RHK6 A0A1Y1NBW2 A0A1Y1NE78 A0A158NAG7 A0A1B6DQ99 A0A1B6E4V7 A0A1B6JE18 A0A1B6GFV2 V5I8J7 A0A1W4WXD7 A0A1B6KPD0 A0A1W4WYN7 A0A1B6KHZ3 A0A1L8DDW9 A0A1L8DDJ4 A0A2L0RID4 A0A2J7PW27 A0A1W4WMQ2 N6UDN4 A0A2M3YZF3 A0A084W8L1 U4U1N4 A0A2S2QYS6 A0A182KA57 A0A182KTL4 A0A182XJL1 A0A182VEC7 A0A1Q3FJZ8 A0A1Q3FJE8 A0A1Q3FJT2 A0A182TW42 A0A182HRI8 Q7PM43 A0A182YR05 A0A1Q3FKZ9 B0WVR2 A0A2C9GL31 A0A139WJW5 A0A1S4JWS2 A0A1B0CTW6 A0A182PCB0 A0A2L0RIE5 A0A0K8TQ57 A0A2H8TWY0 A0A182LX02 A0A182QCM2 A0A224XBK8 A0A0V0G3I9 A0A182RR74 A0A2M4CTL0 A0A2M4CTK4 W5JKP5 A0A2M4CTU7 A0A069DU01 A0A182NAR7 A0A182J8J2 A0A182W913 A0A2M4A4A2 A0A0A9YUE0 A0A3L8D5L8 T1EAB7 A0A182F3V1 A0A0P4VW95 T1E7Y8 A0A0A9YVX6 T1IBH5 A0A2M4A3X2 A0A0K8SD30 A0A182H342 A0A1Y0AWB0 A0A2M4BM04 Q173C1 A0A2M4BL63 A0A1S4FFU4
Pubmed
EMBL
BABH01017075
BABH01017076
GAIX01005381
JAA87179.1
JTDY01006918
KOB65594.1
+ More
GDQN01009486 GDQN01003405 JAT81568.1 JAT87649.1 AGBW02014274 OWR41976.1 ODYU01001598 SOQ38044.1 KQ459605 KPI91878.1 KQ461196 KPJ06418.1 GL888102 EGI67481.1 KQ978623 KYN29721.1 KQ976532 KYM81520.1 KQ434869 KZC09306.1 KQ981463 KYN41501.1 KQ982557 KYQ55057.1 KQ977481 KYN02380.1 GL448571 EFN84148.1 AAZX01012567 JR040212 AEY59033.1 KZ288238 PBC31360.1 KQ435703 KOX80324.1 GL442298 EFN63610.1 KQ766578 OAD53684.1 GL762906 EFZ20489.1 KQ414579 KOC71141.1 LJIG01001072 KRT85868.1 GBYB01012682 JAG82449.1 GEZM01007460 JAV95179.1 GEZM01007459 JAV95180.1 ADTU01000001 GEDC01009434 JAS27864.1 GEDC01004350 JAS32948.1 GECU01010339 JAS97367.1 GECZ01008444 JAS61325.1 GALX01004565 JAB63901.1 GEBQ01026668 JAT13309.1 GEBQ01028900 JAT11077.1 GFDF01009554 JAV04530.1 GFDF01009553 JAV04531.1 KY448282 AUZ41739.1 NEVH01020938 PNF20537.1 APGK01026501 KB740635 ENN79815.1 GGFM01000847 MBW21598.1 ATLV01021492 ATLV01021493 KE525319 KFB46555.1 KB631992 ERL87779.1 GGMS01013682 MBY82885.1 GFDL01007187 JAV27858.1 GFDL01007338 JAV27707.1 GFDL01007257 JAV27788.1 APCN01001708 AAAB01008980 EAA14383.5 GFDL01006817 JAV28228.1 DS232131 EDS35669.1 KQ971329 KYB28338.1 AJWK01028053 KY448285 AUZ41742.1 GDAI01001325 JAI16278.1 GFXV01006496 MBW18301.1 AXCM01000562 AXCN02000660 GFTR01006620 JAW09806.1 GECL01003464 JAP02660.1 GGFL01004506 MBW68684.1 GGFL01004508 MBW68686.1 ADMH02000832 ETN64927.1 GGFL01004507 MBW68685.1 GBGD01001301 JAC87588.1 GGFK01002239 MBW35560.1 GBHO01008358 GDHC01000682 JAG35246.1 JAQ17947.1 QOIP01000012 RLU15787.1 GAMD01000587 JAB01004.1 GDKW01001632 JAI54963.1 GAMD01002647 JAA98943.1 GBHO01008356 JAG35248.1 ACPB03000224 GGFK01002176 MBW35497.1 GBRD01014618 GBRD01014617 JAG51208.1 JXUM01024141 KQ560681 KXJ81292.1 KY921793 ART29382.1 GGFJ01004667 MBW53808.1 CH477427 EAT41139.1 GGFJ01004666 MBW53807.1
GDQN01009486 GDQN01003405 JAT81568.1 JAT87649.1 AGBW02014274 OWR41976.1 ODYU01001598 SOQ38044.1 KQ459605 KPI91878.1 KQ461196 KPJ06418.1 GL888102 EGI67481.1 KQ978623 KYN29721.1 KQ976532 KYM81520.1 KQ434869 KZC09306.1 KQ981463 KYN41501.1 KQ982557 KYQ55057.1 KQ977481 KYN02380.1 GL448571 EFN84148.1 AAZX01012567 JR040212 AEY59033.1 KZ288238 PBC31360.1 KQ435703 KOX80324.1 GL442298 EFN63610.1 KQ766578 OAD53684.1 GL762906 EFZ20489.1 KQ414579 KOC71141.1 LJIG01001072 KRT85868.1 GBYB01012682 JAG82449.1 GEZM01007460 JAV95179.1 GEZM01007459 JAV95180.1 ADTU01000001 GEDC01009434 JAS27864.1 GEDC01004350 JAS32948.1 GECU01010339 JAS97367.1 GECZ01008444 JAS61325.1 GALX01004565 JAB63901.1 GEBQ01026668 JAT13309.1 GEBQ01028900 JAT11077.1 GFDF01009554 JAV04530.1 GFDF01009553 JAV04531.1 KY448282 AUZ41739.1 NEVH01020938 PNF20537.1 APGK01026501 KB740635 ENN79815.1 GGFM01000847 MBW21598.1 ATLV01021492 ATLV01021493 KE525319 KFB46555.1 KB631992 ERL87779.1 GGMS01013682 MBY82885.1 GFDL01007187 JAV27858.1 GFDL01007338 JAV27707.1 GFDL01007257 JAV27788.1 APCN01001708 AAAB01008980 EAA14383.5 GFDL01006817 JAV28228.1 DS232131 EDS35669.1 KQ971329 KYB28338.1 AJWK01028053 KY448285 AUZ41742.1 GDAI01001325 JAI16278.1 GFXV01006496 MBW18301.1 AXCM01000562 AXCN02000660 GFTR01006620 JAW09806.1 GECL01003464 JAP02660.1 GGFL01004506 MBW68684.1 GGFL01004508 MBW68686.1 ADMH02000832 ETN64927.1 GGFL01004507 MBW68685.1 GBGD01001301 JAC87588.1 GGFK01002239 MBW35560.1 GBHO01008358 GDHC01000682 JAG35246.1 JAQ17947.1 QOIP01000012 RLU15787.1 GAMD01000587 JAB01004.1 GDKW01001632 JAI54963.1 GAMD01002647 JAA98943.1 GBHO01008356 JAG35248.1 ACPB03000224 GGFK01002176 MBW35497.1 GBRD01014618 GBRD01014617 JAG51208.1 JXUM01024141 KQ560681 KXJ81292.1 KY921793 ART29382.1 GGFJ01004667 MBW53808.1 CH477427 EAT41139.1 GGFJ01004666 MBW53807.1
Proteomes
UP000005204
UP000037510
UP000007151
UP000053268
UP000053240
UP000007755
+ More
UP000078492 UP000078540 UP000076502 UP000078541 UP000075809 UP000078542 UP000008237 UP000005203 UP000002358 UP000242457 UP000053105 UP000000311 UP000053825 UP000005205 UP000192223 UP000235965 UP000019118 UP000030765 UP000030742 UP000075881 UP000075882 UP000076407 UP000075903 UP000075902 UP000075840 UP000007062 UP000076408 UP000002320 UP000007266 UP000092461 UP000075885 UP000075883 UP000075886 UP000075900 UP000000673 UP000075884 UP000075880 UP000075920 UP000279307 UP000069272 UP000015103 UP000069940 UP000249989 UP000008820
UP000078492 UP000078540 UP000076502 UP000078541 UP000075809 UP000078542 UP000008237 UP000005203 UP000002358 UP000242457 UP000053105 UP000000311 UP000053825 UP000005205 UP000192223 UP000235965 UP000019118 UP000030765 UP000030742 UP000075881 UP000075882 UP000076407 UP000075903 UP000075902 UP000075840 UP000007062 UP000076408 UP000002320 UP000007266 UP000092461 UP000075885 UP000075883 UP000075886 UP000075900 UP000000673 UP000075884 UP000075880 UP000075920 UP000279307 UP000069272 UP000015103 UP000069940 UP000249989 UP000008820
PRIDE
Pfam
PF01490 Aa_trans
Interpro
IPR013057
AA_transpt_TM
ProteinModelPortal
H9JS18
S4P9N7
A0A0L7KRE4
A0A1E1WKV8
A0A212EKG6
A0A2H1VCT6
+ More
A0A194PFS3 A0A194QLI2 F4WE43 A0A151JRF8 A0A195BBC7 A0A154PBN8 A0A151JYP8 A0A151X3R2 A0A195CP10 E2BJJ1 A0A088AJT7 K7JB18 V9ID24 A0A2A3EI11 A0A0N0U7Q8 E2ASE4 A0A310S756 E9IG15 A0A0L7RK76 A0A0T6BEU7 A0A0C9RHK6 A0A1Y1NBW2 A0A1Y1NE78 A0A158NAG7 A0A1B6DQ99 A0A1B6E4V7 A0A1B6JE18 A0A1B6GFV2 V5I8J7 A0A1W4WXD7 A0A1B6KPD0 A0A1W4WYN7 A0A1B6KHZ3 A0A1L8DDW9 A0A1L8DDJ4 A0A2L0RID4 A0A2J7PW27 A0A1W4WMQ2 N6UDN4 A0A2M3YZF3 A0A084W8L1 U4U1N4 A0A2S2QYS6 A0A182KA57 A0A182KTL4 A0A182XJL1 A0A182VEC7 A0A1Q3FJZ8 A0A1Q3FJE8 A0A1Q3FJT2 A0A182TW42 A0A182HRI8 Q7PM43 A0A182YR05 A0A1Q3FKZ9 B0WVR2 A0A2C9GL31 A0A139WJW5 A0A1S4JWS2 A0A1B0CTW6 A0A182PCB0 A0A2L0RIE5 A0A0K8TQ57 A0A2H8TWY0 A0A182LX02 A0A182QCM2 A0A224XBK8 A0A0V0G3I9 A0A182RR74 A0A2M4CTL0 A0A2M4CTK4 W5JKP5 A0A2M4CTU7 A0A069DU01 A0A182NAR7 A0A182J8J2 A0A182W913 A0A2M4A4A2 A0A0A9YUE0 A0A3L8D5L8 T1EAB7 A0A182F3V1 A0A0P4VW95 T1E7Y8 A0A0A9YVX6 T1IBH5 A0A2M4A3X2 A0A0K8SD30 A0A182H342 A0A1Y0AWB0 A0A2M4BM04 Q173C1 A0A2M4BL63 A0A1S4FFU4
A0A194PFS3 A0A194QLI2 F4WE43 A0A151JRF8 A0A195BBC7 A0A154PBN8 A0A151JYP8 A0A151X3R2 A0A195CP10 E2BJJ1 A0A088AJT7 K7JB18 V9ID24 A0A2A3EI11 A0A0N0U7Q8 E2ASE4 A0A310S756 E9IG15 A0A0L7RK76 A0A0T6BEU7 A0A0C9RHK6 A0A1Y1NBW2 A0A1Y1NE78 A0A158NAG7 A0A1B6DQ99 A0A1B6E4V7 A0A1B6JE18 A0A1B6GFV2 V5I8J7 A0A1W4WXD7 A0A1B6KPD0 A0A1W4WYN7 A0A1B6KHZ3 A0A1L8DDW9 A0A1L8DDJ4 A0A2L0RID4 A0A2J7PW27 A0A1W4WMQ2 N6UDN4 A0A2M3YZF3 A0A084W8L1 U4U1N4 A0A2S2QYS6 A0A182KA57 A0A182KTL4 A0A182XJL1 A0A182VEC7 A0A1Q3FJZ8 A0A1Q3FJE8 A0A1Q3FJT2 A0A182TW42 A0A182HRI8 Q7PM43 A0A182YR05 A0A1Q3FKZ9 B0WVR2 A0A2C9GL31 A0A139WJW5 A0A1S4JWS2 A0A1B0CTW6 A0A182PCB0 A0A2L0RIE5 A0A0K8TQ57 A0A2H8TWY0 A0A182LX02 A0A182QCM2 A0A224XBK8 A0A0V0G3I9 A0A182RR74 A0A2M4CTL0 A0A2M4CTK4 W5JKP5 A0A2M4CTU7 A0A069DU01 A0A182NAR7 A0A182J8J2 A0A182W913 A0A2M4A4A2 A0A0A9YUE0 A0A3L8D5L8 T1EAB7 A0A182F3V1 A0A0P4VW95 T1E7Y8 A0A0A9YVX6 T1IBH5 A0A2M4A3X2 A0A0K8SD30 A0A182H342 A0A1Y0AWB0 A0A2M4BM04 Q173C1 A0A2M4BL63 A0A1S4FFU4
Ontologies
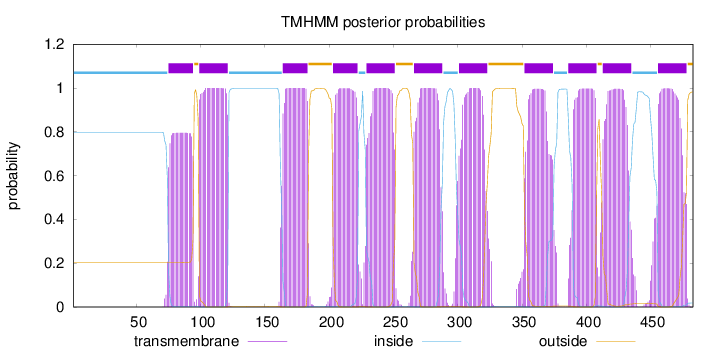
Topology
Length:
483
Number of predicted TMHs:
11
Exp number of AAs in TMHs:
232.67926
Exp number, first 60 AAs:
0
Total prob of N-in:
0.79696
inside
1 - 74
TMhelix
75 - 94
outside
95 - 98
TMhelix
99 - 121
inside
122 - 163
TMhelix
164 - 183
outside
184 - 202
TMhelix
203 - 222
inside
223 - 228
TMhelix
229 - 251
outside
252 - 265
TMhelix
266 - 288
inside
289 - 300
TMhelix
301 - 323
outside
324 - 351
TMhelix
352 - 374
inside
375 - 385
TMhelix
386 - 408
outside
409 - 412
TMhelix
413 - 435
inside
436 - 455
TMhelix
456 - 478
outside
479 - 483
Population Genetic Test Statistics
Pi
22.932232
Theta
21.154437
Tajima's D
-1.539018
CLR
0.021497
CSRT
0.0542972851357432
Interpretation
Possibly Positive selection
