Gene
KWMTBOMO00534
Pre Gene Modal
BGIBMGA012266
Annotation
PREDICTED:_glucuronyltransferase_isoform_X1_[Bombyx_mori]
Full name
Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase
+ More
Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase S
Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase S
Alternative Name
Beta-1,3-glucuronyltransferase S
Glucuronosyltransferase S
UDP-glucuronosyltransferase S
Glucuronosyltransferase S
UDP-glucuronosyltransferase S
Location in the cell
PlasmaMembrane Reliability : 2.542
Sequence
CDS
ATGAAATCAATTTGTTTACTTAAGAAAGTTTTTGTGATGGCTATATTCCTTTGTACATTTTTCTTAGTGTTTTTCTGGGCAAATATGCAAATTTTAAACTTGAATGGAGTAGACACCGCCCTGATATCAAAAATGAGGCCACTGCATGTGAAGAGCAGATTGTGTCATGTTAATTTAATTGACGGCAGAAATCATAACGAAGCTATAGCGAATAAGAGCGATTTGAAAATGATATATTACATCACGCCGACTTATCCAAGACCAGAACAGATTCCAGAGTTAACTAGGCTTGGCCACACATTGATGCACGTGCCTCGGATACATTGGATCATTGCAGACGACCAGTCTTTGTGTTCAACCAATGTTCTCAATTTACTTAGGCGGACGGGCTTGCCATTCACACATATTTCCAGTCCAAAGCCATACGTGTACAAAGGTACAAACTTCCCGCGTGGCGTTTCGAACAGACGTGCAGCCCTGGTTTGGTTGCGGGAGAACGTTCGAGAAGGAGTCATGTACTTTGGCGATGACGATAATACAGTAGACTTACAGCTATTCGATGAAATAAGGCGTACCAAGAAGGTATCCATGTTTCCCGTAGGGCTCATTGGTGATTACGGTATTTCGGCGCCCATCATCAAAGACGGAAAGGTCGTCGCCTTTTTCGACTCATGGCCGGGGTCTAGAACATTCCCTGTCGATATGGCAGGATTCGCTGTCAACATAGAGTTCCTGACTCCGACGGCCACAATGCCTTACTCAGCTGGCCACGAGGAAGATAAGTTTTTAATGAGTTTAGGAATAAAATTAGACGACATTGAACCGTTAGCCGATAATTGTTCAAAGGTTTTAGTTTGGCACACGAAAACTGTAAAGTTTAAAAAACCCAACCTTAAAATCGATATCGAAAGAATTAGCAAGTTACCGAAGTACGAGGATTTCGCTAATCTTTTAAAAGAAACTTCTAGGTTAGGTATGGCCGGAATATTCTCGAAAAATGGTAGTAAAACTTTCATAATTAAAGATAGGAAGACCTTTGAACCCTTAAGCCTTCTGCCCACTTTGGTATAA
Protein
MKSICLLKKVFVMAIFLCTFFLVFFWANMQILNLNGVDTALISKMRPLHVKSRLCHVNLIDGRNHNEAIANKSDLKMIYYITPTYPRPEQIPELTRLGHTLMHVPRIHWIIADDQSLCSTNVLNLLRRTGLPFTHISSPKPYVYKGTNFPRGVSNRRAALVWLRENVREGVMYFGDDDNTVDLQLFDEIRRTKKVSMFPVGLIGDYGISAPIIKDGKVVAFFDSWPGSRTFPVDMAGFAVNIEFLTPTATMPYSAGHEEDKFLMSLGIKLDDIEPLADNCSKVLVWHTKTVKFKKPNLKIDIERISKLPKYEDFANLLKETSRLGMAGIFSKNGSKTFIIKDRKTFEPLSLLPTLV
Summary
Description
Involved in the biosynthesis of L2/HNK-1 carbohydrate epitope on both glycolipids and glycoproteins. Enzyme has a broad specificity.
Catalytic Activity
3-O-(beta-D-galactosyl-(1->3)-beta-D-galactosyl-(1->4)-beta-D-xylosyl)-L-seryl-[protein] + UDP-alpha-D-glucuronate = 3-O-(beta-D-GlcA-(1->3)-beta-D-Gal-(1->3)-beta-D-Gal-(1->4)-beta-D-Xyl)-L-seryl-[protein] + H(+) + UDP
Cofactor
Mn(2+)
Similarity
Belongs to the glycosyltransferase 43 family.
Keywords
Alternative splicing
Complete proteome
Glycoprotein
Golgi apparatus
Manganese
Membrane
Metal-binding
Phosphoprotein
Reference proteome
Signal-anchor
Transferase
Transmembrane
Transmembrane helix
Feature
chain Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase
splice variant In isoform a.
splice variant In isoform a.
Uniprot
H9JRV6
B5A9M8
A0A2A4J695
A0A194QUI9
A0A1E1WHQ8
A0A2H1V9Z4
+ More
A0A212EKH4 A0A194PET1 A0A067RD25 A0A2P8Z5Y6 A0A2J7PNA8 A0A1B6LFL3 A0A1S3CUY6 A0A1B6EZJ1 A0A1B6BXY2 A0A1B0C836 A0A1L8E213 A0A1Q3F083 Q17BE2 A0A1Q3F0X4 A0A182KEA7 A0A1Q3F126 A0A182IPX0 A0A182M4K6 A0A1Q3EZJ8 A0A1Q3F1I9 A0A1Q3F075 A0A336LV95 A0A182QZC4 A0A182V4K2 A0A182X609 A0A084VJ92 A0A182U069 A0A182R2J4 A0A1Y9GKD1 A0A1S4GZB1 Q17BE4 A0A182IB33 A0A2M4AY11 B0WPV1 U5EGY0 A0A2M3ZBP9 A0A182PTY9 A0A182NT56 A0A2M3ZBK1 A0A182WA06 A0A2S2QEC9 A0A2M4BR33 A0A2H8TJQ4 A0A1J1IG96 A0A2M4CQ49 W5JCX8 Q5CAS4 J9JQS1 A0A1A9V9N1 A0A1B0A4Y3 A0A2S2P568 Q29NA9 A0A0R3NRT8 A0A1W4VHW5 A0A1W4VVE3 A0A1W4VUM4 A0A1W4W2A1 A0A1I8P808 A0A0K8UDI5 A0A1W4VW77 B3MUX2 A0A1A9W007 A0A1I8MSQ4 Q5CAS3 A0A3B0JX87 A0A1A9XGW8 A0A1B0BUW9 B4NY61 A0A3B0JUA3 A0A1I8MSQ6 T1PHC2 B4Q7U6 C6SUZ1 A0A182G278 Q9VLA1-2 A0A1I8P852 C0PTW3 B3N8G2 A0A2Z5Y5E9 Q9VLA1 A0A1B0GQ87 E1JHC8 A0A0R1DS95 A0A0P8XXU3 A0A3B0JUA0 B4HW83 A0A0J9TJL6 A0A0A1XBQ6 A0A0M3QTK1 W8BLI9 A0A0L0BL08
A0A212EKH4 A0A194PET1 A0A067RD25 A0A2P8Z5Y6 A0A2J7PNA8 A0A1B6LFL3 A0A1S3CUY6 A0A1B6EZJ1 A0A1B6BXY2 A0A1B0C836 A0A1L8E213 A0A1Q3F083 Q17BE2 A0A1Q3F0X4 A0A182KEA7 A0A1Q3F126 A0A182IPX0 A0A182M4K6 A0A1Q3EZJ8 A0A1Q3F1I9 A0A1Q3F075 A0A336LV95 A0A182QZC4 A0A182V4K2 A0A182X609 A0A084VJ92 A0A182U069 A0A182R2J4 A0A1Y9GKD1 A0A1S4GZB1 Q17BE4 A0A182IB33 A0A2M4AY11 B0WPV1 U5EGY0 A0A2M3ZBP9 A0A182PTY9 A0A182NT56 A0A2M3ZBK1 A0A182WA06 A0A2S2QEC9 A0A2M4BR33 A0A2H8TJQ4 A0A1J1IG96 A0A2M4CQ49 W5JCX8 Q5CAS4 J9JQS1 A0A1A9V9N1 A0A1B0A4Y3 A0A2S2P568 Q29NA9 A0A0R3NRT8 A0A1W4VHW5 A0A1W4VVE3 A0A1W4VUM4 A0A1W4W2A1 A0A1I8P808 A0A0K8UDI5 A0A1W4VW77 B3MUX2 A0A1A9W007 A0A1I8MSQ4 Q5CAS3 A0A3B0JX87 A0A1A9XGW8 A0A1B0BUW9 B4NY61 A0A3B0JUA3 A0A1I8MSQ6 T1PHC2 B4Q7U6 C6SUZ1 A0A182G278 Q9VLA1-2 A0A1I8P852 C0PTW3 B3N8G2 A0A2Z5Y5E9 Q9VLA1 A0A1B0GQ87 E1JHC8 A0A0R1DS95 A0A0P8XXU3 A0A3B0JUA0 B4HW83 A0A0J9TJL6 A0A0A1XBQ6 A0A0M3QTK1 W8BLI9 A0A0L0BL08
EC Number
2.4.1.135
Pubmed
19121390
26354079
22118469
24845553
29403074
17510324
+ More
24438588 12364791 20920257 23761445 15632085 17994087 25315136 17550304 26483478 10731132 12537572 12537569 12511570 18327897 29499182 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25830018 24495485 26108605
24438588 12364791 20920257 23761445 15632085 17994087 25315136 17550304 26483478 10731132 12537572 12537569 12511570 18327897 29499182 12537568 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25830018 24495485 26108605
EMBL
BABH01017062
BABH01017063
EU827503
ACF31554.2
NWSH01002712
PCG67637.1
+ More
KQ461108 KPJ08989.1 GDQN01004618 JAT86436.1 ODYU01001436 SOQ37621.1 AGBW02014274 OWR41984.1 KQ459605 KPI91871.1 KK852715 KDR17861.1 PYGN01000181 PSN51915.1 NEVH01023955 PNF17820.1 GEBQ01017623 JAT22354.1 GECZ01026293 JAS43476.1 GEDC01031156 JAS06142.1 AJWK01000126 GFDF01001326 JAV12758.1 GFDL01014083 JAV20962.1 CH477322 EAT43601.1 GFDL01013843 JAV21202.1 GFDL01013857 JAV21188.1 AXCM01000090 GFDL01014318 JAV20727.1 GFDL01013630 JAV21415.1 GFDL01014080 JAV20965.1 UFQT01000114 SSX20257.1 AXCN02000729 ATLV01013476 KE524860 KFB38036.1 APCN01000637 AAAB01008944 EAT43602.1 GGFK01012314 MBW45635.1 DS232030 EDS32539.1 GANO01003250 JAB56621.1 GGFM01005139 MBW25890.1 GGFM01005183 MBW25934.1 GGMS01006891 MBY76094.1 GGFJ01006378 MBW55519.1 GFXV01002406 MBW14211.1 CVRI01000047 CRK98564.1 GGFL01003133 MBW67311.1 ADMH02001599 ETN61901.1 AJ889609 CAI63867.1 ABLF02019600 ABLF02019602 GGMR01012022 MBY24641.1 CH379060 EAL33433.2 KRT03826.1 GDHF01027615 JAI24699.1 CH902624 EDV33037.2 AJ889610 CAI63868.1 OUUW01000010 SPP85683.1 JXJN01020958 CM000157 EDW88663.1 SPP85684.1 KA648227 AFP62856.1 CM000361 EDX04378.1 BT088983 ACT88124.1 JXUM01039461 JXUM01039462 JXUM01039463 JXUM01039464 JXUM01039465 JXUM01039466 JXUM01039467 KQ561205 KXJ79350.1 AE014134 AY058399 AB080696 AAF52795.2 BT071808 ACN58571.1 CH954177 EDV58385.1 LC146675 BBC62124.1 AJVK01006586 AJVK01006587 BT133218 ACZ94212.1 AFA28459.1 KRJ97968.1 KPU74267.1 SPP85685.1 CH480818 EDW52278.1 CM002910 KMY89295.1 GBXI01011999 GBXI01005433 JAD02293.1 JAD08859.1 CP012523 ALC39053.1 GAMC01006883 JAB99672.1 JRES01001704 KNC20732.1
KQ461108 KPJ08989.1 GDQN01004618 JAT86436.1 ODYU01001436 SOQ37621.1 AGBW02014274 OWR41984.1 KQ459605 KPI91871.1 KK852715 KDR17861.1 PYGN01000181 PSN51915.1 NEVH01023955 PNF17820.1 GEBQ01017623 JAT22354.1 GECZ01026293 JAS43476.1 GEDC01031156 JAS06142.1 AJWK01000126 GFDF01001326 JAV12758.1 GFDL01014083 JAV20962.1 CH477322 EAT43601.1 GFDL01013843 JAV21202.1 GFDL01013857 JAV21188.1 AXCM01000090 GFDL01014318 JAV20727.1 GFDL01013630 JAV21415.1 GFDL01014080 JAV20965.1 UFQT01000114 SSX20257.1 AXCN02000729 ATLV01013476 KE524860 KFB38036.1 APCN01000637 AAAB01008944 EAT43602.1 GGFK01012314 MBW45635.1 DS232030 EDS32539.1 GANO01003250 JAB56621.1 GGFM01005139 MBW25890.1 GGFM01005183 MBW25934.1 GGMS01006891 MBY76094.1 GGFJ01006378 MBW55519.1 GFXV01002406 MBW14211.1 CVRI01000047 CRK98564.1 GGFL01003133 MBW67311.1 ADMH02001599 ETN61901.1 AJ889609 CAI63867.1 ABLF02019600 ABLF02019602 GGMR01012022 MBY24641.1 CH379060 EAL33433.2 KRT03826.1 GDHF01027615 JAI24699.1 CH902624 EDV33037.2 AJ889610 CAI63868.1 OUUW01000010 SPP85683.1 JXJN01020958 CM000157 EDW88663.1 SPP85684.1 KA648227 AFP62856.1 CM000361 EDX04378.1 BT088983 ACT88124.1 JXUM01039461 JXUM01039462 JXUM01039463 JXUM01039464 JXUM01039465 JXUM01039466 JXUM01039467 KQ561205 KXJ79350.1 AE014134 AY058399 AB080696 AAF52795.2 BT071808 ACN58571.1 CH954177 EDV58385.1 LC146675 BBC62124.1 AJVK01006586 AJVK01006587 BT133218 ACZ94212.1 AFA28459.1 KRJ97968.1 KPU74267.1 SPP85685.1 CH480818 EDW52278.1 CM002910 KMY89295.1 GBXI01011999 GBXI01005433 JAD02293.1 JAD08859.1 CP012523 ALC39053.1 GAMC01006883 JAB99672.1 JRES01001704 KNC20732.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000027135
+ More
UP000245037 UP000235965 UP000079169 UP000092461 UP000008820 UP000075881 UP000075880 UP000075883 UP000075886 UP000075903 UP000076407 UP000030765 UP000075902 UP000075900 UP000075840 UP000002320 UP000075885 UP000075884 UP000075920 UP000183832 UP000000673 UP000007819 UP000078200 UP000092445 UP000001819 UP000192221 UP000095300 UP000007801 UP000091820 UP000095301 UP000268350 UP000092443 UP000092460 UP000002282 UP000000304 UP000069940 UP000249989 UP000000803 UP000008711 UP000092462 UP000001292 UP000092553 UP000037069
UP000245037 UP000235965 UP000079169 UP000092461 UP000008820 UP000075881 UP000075880 UP000075883 UP000075886 UP000075903 UP000076407 UP000030765 UP000075902 UP000075900 UP000075840 UP000002320 UP000075885 UP000075884 UP000075920 UP000183832 UP000000673 UP000007819 UP000078200 UP000092445 UP000001819 UP000192221 UP000095300 UP000007801 UP000091820 UP000095301 UP000268350 UP000092443 UP000092460 UP000002282 UP000000304 UP000069940 UP000249989 UP000000803 UP000008711 UP000092462 UP000001292 UP000092553 UP000037069
Interpro
Gene 3D
CDD
ProteinModelPortal
H9JRV6
B5A9M8
A0A2A4J695
A0A194QUI9
A0A1E1WHQ8
A0A2H1V9Z4
+ More
A0A212EKH4 A0A194PET1 A0A067RD25 A0A2P8Z5Y6 A0A2J7PNA8 A0A1B6LFL3 A0A1S3CUY6 A0A1B6EZJ1 A0A1B6BXY2 A0A1B0C836 A0A1L8E213 A0A1Q3F083 Q17BE2 A0A1Q3F0X4 A0A182KEA7 A0A1Q3F126 A0A182IPX0 A0A182M4K6 A0A1Q3EZJ8 A0A1Q3F1I9 A0A1Q3F075 A0A336LV95 A0A182QZC4 A0A182V4K2 A0A182X609 A0A084VJ92 A0A182U069 A0A182R2J4 A0A1Y9GKD1 A0A1S4GZB1 Q17BE4 A0A182IB33 A0A2M4AY11 B0WPV1 U5EGY0 A0A2M3ZBP9 A0A182PTY9 A0A182NT56 A0A2M3ZBK1 A0A182WA06 A0A2S2QEC9 A0A2M4BR33 A0A2H8TJQ4 A0A1J1IG96 A0A2M4CQ49 W5JCX8 Q5CAS4 J9JQS1 A0A1A9V9N1 A0A1B0A4Y3 A0A2S2P568 Q29NA9 A0A0R3NRT8 A0A1W4VHW5 A0A1W4VVE3 A0A1W4VUM4 A0A1W4W2A1 A0A1I8P808 A0A0K8UDI5 A0A1W4VW77 B3MUX2 A0A1A9W007 A0A1I8MSQ4 Q5CAS3 A0A3B0JX87 A0A1A9XGW8 A0A1B0BUW9 B4NY61 A0A3B0JUA3 A0A1I8MSQ6 T1PHC2 B4Q7U6 C6SUZ1 A0A182G278 Q9VLA1-2 A0A1I8P852 C0PTW3 B3N8G2 A0A2Z5Y5E9 Q9VLA1 A0A1B0GQ87 E1JHC8 A0A0R1DS95 A0A0P8XXU3 A0A3B0JUA0 B4HW83 A0A0J9TJL6 A0A0A1XBQ6 A0A0M3QTK1 W8BLI9 A0A0L0BL08
A0A212EKH4 A0A194PET1 A0A067RD25 A0A2P8Z5Y6 A0A2J7PNA8 A0A1B6LFL3 A0A1S3CUY6 A0A1B6EZJ1 A0A1B6BXY2 A0A1B0C836 A0A1L8E213 A0A1Q3F083 Q17BE2 A0A1Q3F0X4 A0A182KEA7 A0A1Q3F126 A0A182IPX0 A0A182M4K6 A0A1Q3EZJ8 A0A1Q3F1I9 A0A1Q3F075 A0A336LV95 A0A182QZC4 A0A182V4K2 A0A182X609 A0A084VJ92 A0A182U069 A0A182R2J4 A0A1Y9GKD1 A0A1S4GZB1 Q17BE4 A0A182IB33 A0A2M4AY11 B0WPV1 U5EGY0 A0A2M3ZBP9 A0A182PTY9 A0A182NT56 A0A2M3ZBK1 A0A182WA06 A0A2S2QEC9 A0A2M4BR33 A0A2H8TJQ4 A0A1J1IG96 A0A2M4CQ49 W5JCX8 Q5CAS4 J9JQS1 A0A1A9V9N1 A0A1B0A4Y3 A0A2S2P568 Q29NA9 A0A0R3NRT8 A0A1W4VHW5 A0A1W4VVE3 A0A1W4VUM4 A0A1W4W2A1 A0A1I8P808 A0A0K8UDI5 A0A1W4VW77 B3MUX2 A0A1A9W007 A0A1I8MSQ4 Q5CAS3 A0A3B0JX87 A0A1A9XGW8 A0A1B0BUW9 B4NY61 A0A3B0JUA3 A0A1I8MSQ6 T1PHC2 B4Q7U6 C6SUZ1 A0A182G278 Q9VLA1-2 A0A1I8P852 C0PTW3 B3N8G2 A0A2Z5Y5E9 Q9VLA1 A0A1B0GQ87 E1JHC8 A0A0R1DS95 A0A0P8XXU3 A0A3B0JUA0 B4HW83 A0A0J9TJL6 A0A0A1XBQ6 A0A0M3QTK1 W8BLI9 A0A0L0BL08
PDB
2D0J
E-value=2.67969e-51,
Score=510
Ontologies
GO
PANTHER
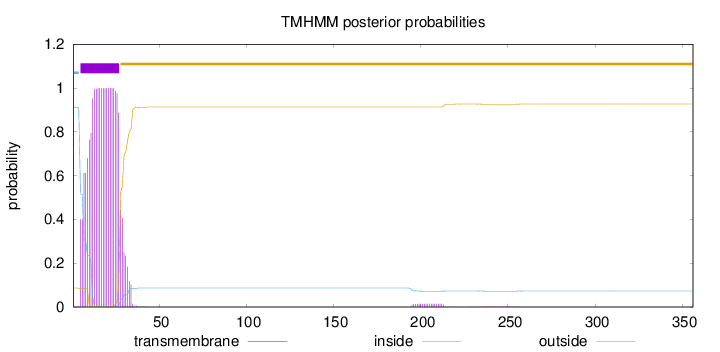
Topology
Subcellular location
Golgi apparatus membrane
Length:
356
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
22.13473
Exp number, first 60 AAs:
21.82198
Total prob of N-in:
0.91317
POSSIBLE N-term signal
sequence
inside
1 - 4
TMhelix
5 - 27
outside
28 - 356
Population Genetic Test Statistics
Pi
2.046429
Theta
13.002571
Tajima's D
-2.162954
CLR
0.174511
CSRT
0.00594970251487426
Interpretation
Uncertain
