Pre Gene Modal
BGIBMGA012325
Annotation
PREDICTED:_ADP-ribosylation_factor-like_protein_5B_[Amyelois_transitella]
Location in the cell
Extracellular Reliability : 1.218
Sequence
CDS
ATGGGCTTACTTATTTCGAAAATATGGAGCTTATTCGGAAATGAGGAACATAAACTCGTATTGGTGGGTCTCGACAATGCTGGCAAAACCACTATTTTATACCAACTACTTCTTGGTGAAGCTGTACATACTCGGCCCACCATAGGGTCGAATGTTGAAGAGGTTGTGTGGAAGAATCTCCGGTTTGTGATGTGGGACTTAGGAGGACAACAAAGCTTGAGATCAGCATGGAATACTTACTACACAAATAGTGAGTTCGTTATAATGGTGATCGACTCGACAGATCGACAGCGTTTAGGCATAAGCCGGGAAGAGCTGCATCGCATACTTGTCCACGAAGAACTAAGCAAGGCATGCTTACTCGTATTCGCTAATAAACAGGACGTTAAAGGATCAATGACAGCAGCTGAAATATCGGAGCAATTAGATTTGACGTCGATAAAGCAACATCCCTGGCATATACAGGCGTGTTGCGCGCTCACTGGTGAGGGGTTACATTTAGGTTTAGAATGGATCGCCAGTCGAATAAAGAAGAAATGA
Protein
MGLLISKIWSLFGNEEHKLVLVGLDNAGKTTILYQLLLGEAVHTRPTIGSNVEEVVWKNLRFVMWDLGGQQSLRSAWNTYYTNSEFVIMVIDSTDRQRLGISREELHRILVHEELSKACLLVFANKQDVKGSMTAAEISEQLDLTSIKQHPWHIQACCALTGEGLHLGLEWIASRIKKK
Summary
Similarity
Belongs to the small GTPase superfamily. Arf family.
Uniprot
A0A212EKG4
A0A194QTY5
I4DP13
I4DS43
A0A1E1W2Y6
A0A2A4JIG8
+ More
A0A2A4JC03 A0A2W1BLV2 H9JS15 A0A2H1V0X6 A0A1I8M4N5 A0A1I8PP48 A0A0L0C999 D3TPP5 A0A0K8TS95 W8BW40 U5EYZ5 A0A0K8V7U6 A0A034VWN4 A0A1B6L840 A0A1B6FID0 A0A1B6JW04 T1DQT4 A0A2M3Z970 A0A2M4AP04 W5J7B8 A0A182FWA7 A0A1Q3F9U3 B0XJN1 Q16I84 A0A023EIZ8 A0A182N343 A0A182QBH0 A0A182VCL7 A0A182WDA5 A0A0A1X4D7 A0A182IMZ1 A0A067QPM5 E2AVF5 A0A3L8D7T2 A0A1L8DVP8 F4W8J0 A0A195BAV4 A0A158NK92 B4L950 A0A0L7KXI9 A0A0P4VRW2 A0A023F9M3 R4G4G9 A0A0J7KSR4 A0A0A9WHZ7 A0A1W4US96 A0A3B0JZM0 Q29F85 B4H156 B4PG53 A0A2A3ELK2 A0A088AKA9 B4HJF7 B3NFU8 B4QLL6 Q9VSG8 A0A1B6CDL7 B4LC98 B4J200 J3JVT0 B3M9D8 A0A0M4EAS7 B4MKS4 A0A224XQU1 A0A069DPW1 D6WN25 A0A164XDF0 A0A1Y1LI04 E9H226 A0A1W4WKE8 A0A1B0FLP3 A0A165VG85 Q7QHM4 A0A232FDY7 N6TKI6 A0A0P5X264 T1JPA3 D1FPN0 A0A1S3KG17 C3YNK8 A0A1S3DIT2 A0A182HG29 A0A182LHI2 A0A2C9JLH4 E2BRV1 A0A2S2NTS4 T1E2H0 R7VLQ6 C4WTJ1 T1G522 Q2XYK6 Q2XYK1 Q2XYK5 Q2XYK3
A0A2A4JC03 A0A2W1BLV2 H9JS15 A0A2H1V0X6 A0A1I8M4N5 A0A1I8PP48 A0A0L0C999 D3TPP5 A0A0K8TS95 W8BW40 U5EYZ5 A0A0K8V7U6 A0A034VWN4 A0A1B6L840 A0A1B6FID0 A0A1B6JW04 T1DQT4 A0A2M3Z970 A0A2M4AP04 W5J7B8 A0A182FWA7 A0A1Q3F9U3 B0XJN1 Q16I84 A0A023EIZ8 A0A182N343 A0A182QBH0 A0A182VCL7 A0A182WDA5 A0A0A1X4D7 A0A182IMZ1 A0A067QPM5 E2AVF5 A0A3L8D7T2 A0A1L8DVP8 F4W8J0 A0A195BAV4 A0A158NK92 B4L950 A0A0L7KXI9 A0A0P4VRW2 A0A023F9M3 R4G4G9 A0A0J7KSR4 A0A0A9WHZ7 A0A1W4US96 A0A3B0JZM0 Q29F85 B4H156 B4PG53 A0A2A3ELK2 A0A088AKA9 B4HJF7 B3NFU8 B4QLL6 Q9VSG8 A0A1B6CDL7 B4LC98 B4J200 J3JVT0 B3M9D8 A0A0M4EAS7 B4MKS4 A0A224XQU1 A0A069DPW1 D6WN25 A0A164XDF0 A0A1Y1LI04 E9H226 A0A1W4WKE8 A0A1B0FLP3 A0A165VG85 Q7QHM4 A0A232FDY7 N6TKI6 A0A0P5X264 T1JPA3 D1FPN0 A0A1S3KG17 C3YNK8 A0A1S3DIT2 A0A182HG29 A0A182LHI2 A0A2C9JLH4 E2BRV1 A0A2S2NTS4 T1E2H0 R7VLQ6 C4WTJ1 T1G522 Q2XYK6 Q2XYK1 Q2XYK5 Q2XYK3
Pubmed
22118469
26354079
22651552
28756777
19121390
25315136
+ More
26108605 20353571 26369729 24495485 25348373 20920257 23761445 17510324 24945155 25830018 24845553 20798317 30249741 21719571 21347285 17994087 26227816 27129103 25474469 25401762 26823975 15632085 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22516182 26334808 18362917 19820115 28004739 21292972 12364791 14747013 17210077 28648823 23537049 20441151 18563158 20966253 15562597 24330624 23254933 16120803 18288436
26108605 20353571 26369729 24495485 25348373 20920257 23761445 17510324 24945155 25830018 24845553 20798317 30249741 21719571 21347285 17994087 26227816 27129103 25474469 25401762 26823975 15632085 17550304 22936249 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22516182 26334808 18362917 19820115 28004739 21292972 12364791 14747013 17210077 28648823 23537049 20441151 18563158 20966253 15562597 24330624 23254933 16120803 18288436
EMBL
AGBW02014274
OWR41989.1
KQ461108
KPJ08993.1
AK403239
KQ459605
+ More
BAM19653.1 KPI91868.1 AK405412 BAM20733.1 GDQN01009716 JAT81338.1 NWSH01001311 PCG71761.1 NWSH01002125 PCG69084.1 KZ149971 PZC76052.1 BABH01017048 ODYU01000165 SOQ34471.1 JRES01000741 KNC28822.1 CCAG010021577 EZ423397 ADD19673.1 GDAI01000374 JAI17229.1 GAMC01009084 GAMC01009083 GAMC01009082 GAMC01009081 JAB97471.1 GANO01001712 JAB58159.1 GDHF01023644 GDHF01017664 GDHF01008035 JAI28670.1 JAI34650.1 JAI44279.1 GAKP01012435 GAKP01012434 GAKP01012433 GAKP01012432 JAC46517.1 GEBQ01020094 JAT19883.1 GECZ01019810 JAS49959.1 GECU01014192 GECU01005626 GECU01004328 JAS93514.1 JAT02081.1 JAT03379.1 GAMD01002274 JAA99316.1 GGFM01004227 MBW24978.1 GGFK01009182 MBW42503.1 ADMH02002028 GGFL01004226 ETN59871.1 MBW68404.1 GFDL01010748 JAV24297.1 DS233564 EDS30550.1 CH478097 EAT33975.1 GAPW01004924 GEHC01000328 JAC08674.1 JAV47317.1 AXCN02001130 GBXI01011909 GBXI01008108 GBXI01002577 JAD02383.1 JAD06184.1 JAD11715.1 KK853090 KDR11571.1 GL443090 EFN62589.1 QOIP01000011 RLU16540.1 GFDF01003570 JAV10514.1 GL887908 EGI69481.1 KQ976537 KYM81350.1 ADTU01018741 CH933816 EDW17225.1 JTDY01004592 KOB67972.1 GDKW01000786 JAI55809.1 GBBI01000511 JAC18201.1 ACPB03001046 GAHY01000781 JAA76729.1 LBMM01003579 KMQ93361.1 GBHO01036573 GBRD01001985 GDHC01014526 JAG07031.1 JAG63836.1 JAQ04103.1 OUUW01000012 SPP87525.1 CH379067 EAL31246.1 CH479202 EDW30033.1 CM000159 EDW93197.1 KZ288217 PBC32374.1 CH480815 EDW40681.1 CH954178 EDV50710.1 CM000363 CM002912 EDX09676.1 KMY98340.1 AE014296 AY095045 AAF50451.1 AAM11373.1 GEDC01026766 GEDC01025759 GEDC01025660 JAS10532.1 JAS11539.1 JAS11638.1 CH940647 EDW68743.1 CH916366 EDV95925.1 BT127348 AEE62310.1 CH902618 EDV41151.1 CP012525 ALC44416.1 CH963847 EDW72780.1 GFTR01002972 JAW13454.1 GBGD01003163 JAC85726.1 KQ971343 EFA04341.1 LRGB01001005 KZS14105.1 GEZM01054989 JAV73274.1 GL732585 EFX74089.1 KT984811 AMZ00363.1 AAAB01008816 EAA05066.1 NNAY01000415 OXU28557.1 APGK01006704 APGK01034104 KB740904 KB736898 KB632006 ENN78433.1 ENN83114.1 ERL87944.1 GDIP01078417 JAM25298.1 JH431954 EZ419780 ACY69953.1 GG666534 EEN58103.1 APCN01005463 GL450042 EFN81584.1 GGMR01007908 MBY20527.1 GALA01000719 JAA94133.1 AMQN01003940 KB292298 ELU17810.1 ABLF02027119 AK340634 BAH71211.1 AMQM01005556 AMQM01005557 KB096983 ESO00273.1 DQ138656 ABA86262.1 DQ138661 ABA86267.1 DQ138657 DQ138658 EU313608 EU313609 EU313610 EU313611 EU313612 EU313613 EU313614 EU313615 EU313616 EU313617 EU313618 EU313619 EU313620 EU313621 EU313622 EU313623 ABA86263.1 ABA86264.1 ABY56408.1 ABY56409.1 ABY56410.1 ABY56411.1 ABY56412.1 ABY56413.1 ABY56414.1 ABY56415.1 ABY56416.1 ABY56417.1 ABY56418.1 ABY56419.1 ABY56420.1 ABY56421.1 ABY56422.1 ABY56423.1 DQ138659 DQ138660 ABA86265.1 ABA86266.1
BAM19653.1 KPI91868.1 AK405412 BAM20733.1 GDQN01009716 JAT81338.1 NWSH01001311 PCG71761.1 NWSH01002125 PCG69084.1 KZ149971 PZC76052.1 BABH01017048 ODYU01000165 SOQ34471.1 JRES01000741 KNC28822.1 CCAG010021577 EZ423397 ADD19673.1 GDAI01000374 JAI17229.1 GAMC01009084 GAMC01009083 GAMC01009082 GAMC01009081 JAB97471.1 GANO01001712 JAB58159.1 GDHF01023644 GDHF01017664 GDHF01008035 JAI28670.1 JAI34650.1 JAI44279.1 GAKP01012435 GAKP01012434 GAKP01012433 GAKP01012432 JAC46517.1 GEBQ01020094 JAT19883.1 GECZ01019810 JAS49959.1 GECU01014192 GECU01005626 GECU01004328 JAS93514.1 JAT02081.1 JAT03379.1 GAMD01002274 JAA99316.1 GGFM01004227 MBW24978.1 GGFK01009182 MBW42503.1 ADMH02002028 GGFL01004226 ETN59871.1 MBW68404.1 GFDL01010748 JAV24297.1 DS233564 EDS30550.1 CH478097 EAT33975.1 GAPW01004924 GEHC01000328 JAC08674.1 JAV47317.1 AXCN02001130 GBXI01011909 GBXI01008108 GBXI01002577 JAD02383.1 JAD06184.1 JAD11715.1 KK853090 KDR11571.1 GL443090 EFN62589.1 QOIP01000011 RLU16540.1 GFDF01003570 JAV10514.1 GL887908 EGI69481.1 KQ976537 KYM81350.1 ADTU01018741 CH933816 EDW17225.1 JTDY01004592 KOB67972.1 GDKW01000786 JAI55809.1 GBBI01000511 JAC18201.1 ACPB03001046 GAHY01000781 JAA76729.1 LBMM01003579 KMQ93361.1 GBHO01036573 GBRD01001985 GDHC01014526 JAG07031.1 JAG63836.1 JAQ04103.1 OUUW01000012 SPP87525.1 CH379067 EAL31246.1 CH479202 EDW30033.1 CM000159 EDW93197.1 KZ288217 PBC32374.1 CH480815 EDW40681.1 CH954178 EDV50710.1 CM000363 CM002912 EDX09676.1 KMY98340.1 AE014296 AY095045 AAF50451.1 AAM11373.1 GEDC01026766 GEDC01025759 GEDC01025660 JAS10532.1 JAS11539.1 JAS11638.1 CH940647 EDW68743.1 CH916366 EDV95925.1 BT127348 AEE62310.1 CH902618 EDV41151.1 CP012525 ALC44416.1 CH963847 EDW72780.1 GFTR01002972 JAW13454.1 GBGD01003163 JAC85726.1 KQ971343 EFA04341.1 LRGB01001005 KZS14105.1 GEZM01054989 JAV73274.1 GL732585 EFX74089.1 KT984811 AMZ00363.1 AAAB01008816 EAA05066.1 NNAY01000415 OXU28557.1 APGK01006704 APGK01034104 KB740904 KB736898 KB632006 ENN78433.1 ENN83114.1 ERL87944.1 GDIP01078417 JAM25298.1 JH431954 EZ419780 ACY69953.1 GG666534 EEN58103.1 APCN01005463 GL450042 EFN81584.1 GGMR01007908 MBY20527.1 GALA01000719 JAA94133.1 AMQN01003940 KB292298 ELU17810.1 ABLF02027119 AK340634 BAH71211.1 AMQM01005556 AMQM01005557 KB096983 ESO00273.1 DQ138656 ABA86262.1 DQ138661 ABA86267.1 DQ138657 DQ138658 EU313608 EU313609 EU313610 EU313611 EU313612 EU313613 EU313614 EU313615 EU313616 EU313617 EU313618 EU313619 EU313620 EU313621 EU313622 EU313623 ABA86263.1 ABA86264.1 ABY56408.1 ABY56409.1 ABY56410.1 ABY56411.1 ABY56412.1 ABY56413.1 ABY56414.1 ABY56415.1 ABY56416.1 ABY56417.1 ABY56418.1 ABY56419.1 ABY56420.1 ABY56421.1 ABY56422.1 ABY56423.1 DQ138659 DQ138660 ABA86265.1 ABA86266.1
Proteomes
UP000007151
UP000053240
UP000053268
UP000218220
UP000005204
UP000095301
+ More
UP000095300 UP000037069 UP000092444 UP000000673 UP000069272 UP000002320 UP000008820 UP000075884 UP000075886 UP000075903 UP000075920 UP000075880 UP000027135 UP000000311 UP000279307 UP000007755 UP000078540 UP000005205 UP000009192 UP000037510 UP000015103 UP000036403 UP000192221 UP000268350 UP000001819 UP000008744 UP000002282 UP000242457 UP000005203 UP000001292 UP000008711 UP000000304 UP000000803 UP000008792 UP000001070 UP000007801 UP000092553 UP000007798 UP000007266 UP000076858 UP000000305 UP000192223 UP000007062 UP000215335 UP000019118 UP000030742 UP000085678 UP000001554 UP000079169 UP000075840 UP000075882 UP000076420 UP000008237 UP000014760 UP000007819 UP000015101
UP000095300 UP000037069 UP000092444 UP000000673 UP000069272 UP000002320 UP000008820 UP000075884 UP000075886 UP000075903 UP000075920 UP000075880 UP000027135 UP000000311 UP000279307 UP000007755 UP000078540 UP000005205 UP000009192 UP000037510 UP000015103 UP000036403 UP000192221 UP000268350 UP000001819 UP000008744 UP000002282 UP000242457 UP000005203 UP000001292 UP000008711 UP000000304 UP000000803 UP000008792 UP000001070 UP000007801 UP000092553 UP000007798 UP000007266 UP000076858 UP000000305 UP000192223 UP000007062 UP000215335 UP000019118 UP000030742 UP000085678 UP000001554 UP000079169 UP000075840 UP000075882 UP000076420 UP000008237 UP000014760 UP000007819 UP000015101
Pfam
PF00025 Arf
SUPFAM
SSF52540
SSF52540
ProteinModelPortal
A0A212EKG4
A0A194QTY5
I4DP13
I4DS43
A0A1E1W2Y6
A0A2A4JIG8
+ More
A0A2A4JC03 A0A2W1BLV2 H9JS15 A0A2H1V0X6 A0A1I8M4N5 A0A1I8PP48 A0A0L0C999 D3TPP5 A0A0K8TS95 W8BW40 U5EYZ5 A0A0K8V7U6 A0A034VWN4 A0A1B6L840 A0A1B6FID0 A0A1B6JW04 T1DQT4 A0A2M3Z970 A0A2M4AP04 W5J7B8 A0A182FWA7 A0A1Q3F9U3 B0XJN1 Q16I84 A0A023EIZ8 A0A182N343 A0A182QBH0 A0A182VCL7 A0A182WDA5 A0A0A1X4D7 A0A182IMZ1 A0A067QPM5 E2AVF5 A0A3L8D7T2 A0A1L8DVP8 F4W8J0 A0A195BAV4 A0A158NK92 B4L950 A0A0L7KXI9 A0A0P4VRW2 A0A023F9M3 R4G4G9 A0A0J7KSR4 A0A0A9WHZ7 A0A1W4US96 A0A3B0JZM0 Q29F85 B4H156 B4PG53 A0A2A3ELK2 A0A088AKA9 B4HJF7 B3NFU8 B4QLL6 Q9VSG8 A0A1B6CDL7 B4LC98 B4J200 J3JVT0 B3M9D8 A0A0M4EAS7 B4MKS4 A0A224XQU1 A0A069DPW1 D6WN25 A0A164XDF0 A0A1Y1LI04 E9H226 A0A1W4WKE8 A0A1B0FLP3 A0A165VG85 Q7QHM4 A0A232FDY7 N6TKI6 A0A0P5X264 T1JPA3 D1FPN0 A0A1S3KG17 C3YNK8 A0A1S3DIT2 A0A182HG29 A0A182LHI2 A0A2C9JLH4 E2BRV1 A0A2S2NTS4 T1E2H0 R7VLQ6 C4WTJ1 T1G522 Q2XYK6 Q2XYK1 Q2XYK5 Q2XYK3
A0A2A4JC03 A0A2W1BLV2 H9JS15 A0A2H1V0X6 A0A1I8M4N5 A0A1I8PP48 A0A0L0C999 D3TPP5 A0A0K8TS95 W8BW40 U5EYZ5 A0A0K8V7U6 A0A034VWN4 A0A1B6L840 A0A1B6FID0 A0A1B6JW04 T1DQT4 A0A2M3Z970 A0A2M4AP04 W5J7B8 A0A182FWA7 A0A1Q3F9U3 B0XJN1 Q16I84 A0A023EIZ8 A0A182N343 A0A182QBH0 A0A182VCL7 A0A182WDA5 A0A0A1X4D7 A0A182IMZ1 A0A067QPM5 E2AVF5 A0A3L8D7T2 A0A1L8DVP8 F4W8J0 A0A195BAV4 A0A158NK92 B4L950 A0A0L7KXI9 A0A0P4VRW2 A0A023F9M3 R4G4G9 A0A0J7KSR4 A0A0A9WHZ7 A0A1W4US96 A0A3B0JZM0 Q29F85 B4H156 B4PG53 A0A2A3ELK2 A0A088AKA9 B4HJF7 B3NFU8 B4QLL6 Q9VSG8 A0A1B6CDL7 B4LC98 B4J200 J3JVT0 B3M9D8 A0A0M4EAS7 B4MKS4 A0A224XQU1 A0A069DPW1 D6WN25 A0A164XDF0 A0A1Y1LI04 E9H226 A0A1W4WKE8 A0A1B0FLP3 A0A165VG85 Q7QHM4 A0A232FDY7 N6TKI6 A0A0P5X264 T1JPA3 D1FPN0 A0A1S3KG17 C3YNK8 A0A1S3DIT2 A0A182HG29 A0A182LHI2 A0A2C9JLH4 E2BRV1 A0A2S2NTS4 T1E2H0 R7VLQ6 C4WTJ1 T1G522 Q2XYK6 Q2XYK1 Q2XYK5 Q2XYK3
PDB
1YZG
E-value=1.40575e-72,
Score=689
Ontologies
GO
Topology
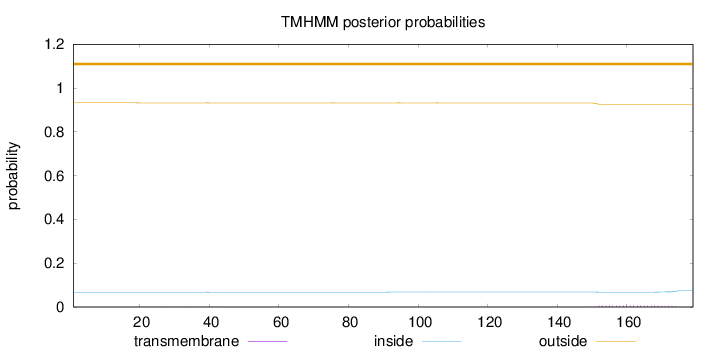
Length:
179
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15264
Exp number, first 60 AAs:
0.01081
Total prob of N-in:
0.06766
outside
1 - 179
Population Genetic Test Statistics
Pi
16.746188
Theta
20.158937
Tajima's D
-1.044451
CLR
0.122477
CSRT
0.130543472826359
Interpretation
Uncertain
