Gene
KWMTBOMO00522
Pre Gene Modal
BGIBMGA012321
Annotation
PREDICTED:_autophagy-related_protein_13_homolog_[Papilio_xuthus]
Full name
Autophagy-related protein 13
+ More
Autophagy-related protein 13 homolog
Autophagy-related protein 13 homolog
Location in the cell
Nuclear Reliability : 3.203
Sequence
CDS
ATGGCAGCATCGAATTCAGAAATTCAAGAACGCTGCCGTTTGTATAAGTTCACCAAAGTTTTGACTTTAAAAGTTGCACAGATTGTGGTCCAGAGCAGGAAAGGAAAGAAAATCAGTCAAGATTTTAAACCACAAAATGGTGTTGATTCTACACCACCTTTGCAATGGTTCAACCTATCAATCCCTGATGAACCTGGAGTTAATGAAGAAACAAAGCGTGTCCTCAATGGAGAAGTAGTTGATGCATTGTCCAAAGTTTTGTCCATAGAAATTACACTTCGAACTACAGATGGTGATGAAATGGTACTTGAATTGTGGACAGTGAAACTGACGACTGCAAGTGAAATATGTTCACCTTCTACAATATACTACAGAATGGGTGTAATGCTTAAGTCGACATTGACAATTTCGCGCATAACCCCAGCATATAAACTTGCACGCACTCAACATAAAGAGACATACAAGATATCCCACCGAATATTTAGCGGAGAACCAAATTTTGAAAAATTGGGAGAGAATCGAAGAAACATCAAAATTAGTGAACTGCGTACACCAATTGGTACAATCACTATTGGTGTATCATACAGGACAAAGATGACAATCTTGCCCGAAGATCGGCCAAAGTTTACATATGGAATAACAACTAGTGGTCATGACATTATGGTTAAGAGTGATCATTTCAAGGATACTCCTAAAAAGAAGAGAGGGTATGAGAAGAAGGAACTTGATCTTTCAAAACCATTAACAAATGCAGCTTTTGTGGATAAAGTTAAAATGCAGGAATTACAGGACTCTTTATGTGAGCAGCTACCACCAGAACCACCGATGACATGGCTCATAGCAGAGAGAGAACAAATCGATAAAAAATTGCAGAATCTCGATATTTGTAAAGTAGCAGATGCGGAGGATCCAACAGCGAAACTTCTAGCGGATGCTGCAAAAACGGCCACGACATCCGCATCAAGCTATGCTGCCAGCAAAGCAATAGATATTCCAAGGGCGAACGCGGGGGATCAGTGCAAGCGCATAATGGAGTTTCCGTTCGCCGAAGGAAGCCCGATCGCGGACCTGGCTAATTTCTACAACGAATGCCTTCAAGCTAGAGCGATGACGAAAGAGTGGAAAGACTTCGCCGATACCGCCGCCATCTCGACCGATTCCGATACTCTTTCCAAGCAGTTGAAGATGTTTGAGGATGCCGTACCGGAATTCGACAGTATGGTCGCTTCAATGTGCAACAGCGCCGAAGGAGGATCCGAGAATTAA
Protein
MAASNSEIQERCRLYKFTKVLTLKVAQIVVQSRKGKKISQDFKPQNGVDSTPPLQWFNLSIPDEPGVNEETKRVLNGEVVDALSKVLSIEITLRTTDGDEMVLELWTVKLTTASEICSPSTIYYRMGVMLKSTLTISRITPAYKLARTQHKETYKISHRIFSGEPNFEKLGENRRNIKISELRTPIGTITIGVSYRTKMTILPEDRPKFTYGITTSGHDIMVKSDHFKDTPKKKRGYEKKELDLSKPLTNAAFVDKVKMQELQDSLCEQLPPEPPMTWLIAEREQIDKKLQNLDICKVADAEDPTAKLLADAAKTATTSASSYAASKAIDIPRANAGDQCKRIMEFPFAEGSPIADLANFYNECLQARAMTKEWKDFADTAAISTDSDTLSKQLKMFEDAVPEFDSMVASMCNSAEGGSEN
Summary
Description
Autophagy factor required for autophagosome formation. Target of the TOR kinase signaling pathway that regulates autophagy through the control of the phosphorylation status of Atg13 and Atg1. The Atg1-Atg13 complex functions at multiple levels to mediate and adjust nutrient-dependent autophagic signaling. Involved in the autophagic degradation of dBruce which controls DNA fragmentation in nurse cells.
Subunit
Interacts with Atg1.
Similarity
Belongs to the ATG13 family. Metazoan subfamily.
Keywords
Autophagy
Complete proteome
Cytoplasm
Phosphoprotein
Reference proteome
Feature
chain Autophagy-related protein 13 homolog
Uniprot
A0A2A4JYG5
A0A2H1W515
A0A194QVK7
A0A194PES2
A0A0L7LDJ7
A0A212EKI8
+ More
A0A1E1WSR5 A0A194QEL5 A0A0N1PIT3 A0A1J1IX18 A0A2H1W5V8 A0A2A4JXB2 A0A067QS73 A0A1E1WL78 A0A2P8Y802 D6WFS6 A0A088AJZ1 A0A2A3EK01 A0A154PSY9 A0A310SEA5 A0A0L7RJK7 A0A232ET24 A0A1L8DV56 A0A1L8DVK6 U5EL05 A0A0A9WT81 A0A195CP19 A0A026VZU1 F4WE30 A0A1Q3G2Y3 A0A1Q3G3G6 A0A151K0C2 A0A146LPQ7 Q177Z0 A0A0H3UAL5 A0A3L8D5G8 A0A0L7KI75 Q9VHR6 A0A2M4CRX2 A0A2M4CRU8 B3P4Y2 A0A1J1J630 A0A2M4BKK7 B4PT54 B4HLK7 A0A2M4A1K5 A0A1W4VHP6 A0A2J7PMP5 A0A212FIG9 A0A0M4EES6 B4QYL3 B3M0F1 A0A0M9AC92 A0A0A1X1I5 T1IPJ1 Q298R4 A0A0K8V099 Q7Q6R5 A0A2Y9D0T3 B4LYY6 B4JV55 E0W0V2 A0A151IR64 A0A0L0C0Y2
A0A1E1WSR5 A0A194QEL5 A0A0N1PIT3 A0A1J1IX18 A0A2H1W5V8 A0A2A4JXB2 A0A067QS73 A0A1E1WL78 A0A2P8Y802 D6WFS6 A0A088AJZ1 A0A2A3EK01 A0A154PSY9 A0A310SEA5 A0A0L7RJK7 A0A232ET24 A0A1L8DV56 A0A1L8DVK6 U5EL05 A0A0A9WT81 A0A195CP19 A0A026VZU1 F4WE30 A0A1Q3G2Y3 A0A1Q3G3G6 A0A151K0C2 A0A146LPQ7 Q177Z0 A0A0H3UAL5 A0A3L8D5G8 A0A0L7KI75 Q9VHR6 A0A2M4CRX2 A0A2M4CRU8 B3P4Y2 A0A1J1J630 A0A2M4BKK7 B4PT54 B4HLK7 A0A2M4A1K5 A0A1W4VHP6 A0A2J7PMP5 A0A212FIG9 A0A0M4EES6 B4QYL3 B3M0F1 A0A0M9AC92 A0A0A1X1I5 T1IPJ1 Q298R4 A0A0K8V099 Q7Q6R5 A0A2Y9D0T3 B4LYY6 B4JV55 E0W0V2 A0A151IR64 A0A0L0C0Y2
Pubmed
EMBL
NWSH01000394
PCG76718.1
ODYU01006344
SOQ48113.1
KQ461108
KPJ09000.1
+ More
KQ459605 KPI91861.1 JTDY01001578 KOB73475.1 AGBW02014274 OWR41996.1 GDQN01000965 JAT90089.1 KQ459054 KPJ03922.1 KQ460047 KPJ17991.1 CVRI01000063 CRL04695.1 ODYU01006546 SOQ48485.1 NWSH01000450 PCG76328.1 KK853018 KDR12447.1 GDQN01003364 GDQN01003281 GDQN01003118 GDQN01000958 JAT87690.1 JAT87773.1 JAT87936.1 JAT90096.1 PYGN01000819 PSN40375.1 KQ971319 EEZ99590.2 KZ288238 PBC31361.1 KQ435191 KZC15035.1 KQ766578 OAD53683.1 KQ414579 KOC71142.1 NNAY01002343 OXU21509.1 GFDF01003760 JAV10324.1 GFDF01003759 JAV10325.1 GANO01001632 JAB58239.1 GBHO01033906 GBRD01009677 JAG09698.1 JAG56147.1 KQ977481 KYN02390.1 KK107558 EZA48976.1 GL888102 EGI67468.1 GFDL01000885 JAV34160.1 GFDL01000729 JAV34316.1 KQ981296 KYN43107.1 GDHC01010012 JAQ08617.1 CH477368 EAT42494.1 KJ778621 AIH07030.1 QOIP01000012 RLU15640.1 JTDY01009683 KOB63047.1 AE014297 AY061109 GGFL01003889 MBW68067.1 GGFL01003888 MBW68066.1 CH954181 EDV49926.1 CVRI01000067 CRL06934.1 GGFJ01004393 MBW53534.1 CM000160 EDW96515.1 CH480815 EDW43034.1 GGFK01001373 MBW34694.1 NEVH01023969 PNF17596.1 AGBW02008391 OWR53518.1 CP012526 ALC46554.1 CM000364 EDX13771.1 CH902617 EDV43157.1 KQ435703 KOX80323.1 GBXI01009667 JAD04625.1 JH431263 CM000070 EAL27891.2 GDHF01019967 JAI32347.1 AAAB01008960 EAA10737.4 APCN01001506 CH940650 EDW68089.1 CH916374 EDV91375.1 DS235862 EEB19258.1 KQ981144 KYN09147.1 JRES01001062 KNC25906.1
KQ459605 KPI91861.1 JTDY01001578 KOB73475.1 AGBW02014274 OWR41996.1 GDQN01000965 JAT90089.1 KQ459054 KPJ03922.1 KQ460047 KPJ17991.1 CVRI01000063 CRL04695.1 ODYU01006546 SOQ48485.1 NWSH01000450 PCG76328.1 KK853018 KDR12447.1 GDQN01003364 GDQN01003281 GDQN01003118 GDQN01000958 JAT87690.1 JAT87773.1 JAT87936.1 JAT90096.1 PYGN01000819 PSN40375.1 KQ971319 EEZ99590.2 KZ288238 PBC31361.1 KQ435191 KZC15035.1 KQ766578 OAD53683.1 KQ414579 KOC71142.1 NNAY01002343 OXU21509.1 GFDF01003760 JAV10324.1 GFDF01003759 JAV10325.1 GANO01001632 JAB58239.1 GBHO01033906 GBRD01009677 JAG09698.1 JAG56147.1 KQ977481 KYN02390.1 KK107558 EZA48976.1 GL888102 EGI67468.1 GFDL01000885 JAV34160.1 GFDL01000729 JAV34316.1 KQ981296 KYN43107.1 GDHC01010012 JAQ08617.1 CH477368 EAT42494.1 KJ778621 AIH07030.1 QOIP01000012 RLU15640.1 JTDY01009683 KOB63047.1 AE014297 AY061109 GGFL01003889 MBW68067.1 GGFL01003888 MBW68066.1 CH954181 EDV49926.1 CVRI01000067 CRL06934.1 GGFJ01004393 MBW53534.1 CM000160 EDW96515.1 CH480815 EDW43034.1 GGFK01001373 MBW34694.1 NEVH01023969 PNF17596.1 AGBW02008391 OWR53518.1 CP012526 ALC46554.1 CM000364 EDX13771.1 CH902617 EDV43157.1 KQ435703 KOX80323.1 GBXI01009667 JAD04625.1 JH431263 CM000070 EAL27891.2 GDHF01019967 JAI32347.1 AAAB01008960 EAA10737.4 APCN01001506 CH940650 EDW68089.1 CH916374 EDV91375.1 DS235862 EEB19258.1 KQ981144 KYN09147.1 JRES01001062 KNC25906.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000037510
UP000007151
UP000183832
+ More
UP000027135 UP000245037 UP000007266 UP000005203 UP000242457 UP000076502 UP000053825 UP000215335 UP000078542 UP000053097 UP000007755 UP000078541 UP000008820 UP000279307 UP000000803 UP000008711 UP000002282 UP000001292 UP000192221 UP000235965 UP000092553 UP000000304 UP000007801 UP000053105 UP000001819 UP000007062 UP000075840 UP000008792 UP000001070 UP000009046 UP000078492 UP000037069
UP000027135 UP000245037 UP000007266 UP000005203 UP000242457 UP000076502 UP000053825 UP000215335 UP000078542 UP000053097 UP000007755 UP000078541 UP000008820 UP000279307 UP000000803 UP000008711 UP000002282 UP000001292 UP000192221 UP000235965 UP000092553 UP000000304 UP000007801 UP000053105 UP000001819 UP000007062 UP000075840 UP000008792 UP000001070 UP000009046 UP000078492 UP000037069
PRIDE
Interpro
SUPFAM
SSF117916
SSF117916
Gene 3D
ProteinModelPortal
A0A2A4JYG5
A0A2H1W515
A0A194QVK7
A0A194PES2
A0A0L7LDJ7
A0A212EKI8
+ More
A0A1E1WSR5 A0A194QEL5 A0A0N1PIT3 A0A1J1IX18 A0A2H1W5V8 A0A2A4JXB2 A0A067QS73 A0A1E1WL78 A0A2P8Y802 D6WFS6 A0A088AJZ1 A0A2A3EK01 A0A154PSY9 A0A310SEA5 A0A0L7RJK7 A0A232ET24 A0A1L8DV56 A0A1L8DVK6 U5EL05 A0A0A9WT81 A0A195CP19 A0A026VZU1 F4WE30 A0A1Q3G2Y3 A0A1Q3G3G6 A0A151K0C2 A0A146LPQ7 Q177Z0 A0A0H3UAL5 A0A3L8D5G8 A0A0L7KI75 Q9VHR6 A0A2M4CRX2 A0A2M4CRU8 B3P4Y2 A0A1J1J630 A0A2M4BKK7 B4PT54 B4HLK7 A0A2M4A1K5 A0A1W4VHP6 A0A2J7PMP5 A0A212FIG9 A0A0M4EES6 B4QYL3 B3M0F1 A0A0M9AC92 A0A0A1X1I5 T1IPJ1 Q298R4 A0A0K8V099 Q7Q6R5 A0A2Y9D0T3 B4LYY6 B4JV55 E0W0V2 A0A151IR64 A0A0L0C0Y2
A0A1E1WSR5 A0A194QEL5 A0A0N1PIT3 A0A1J1IX18 A0A2H1W5V8 A0A2A4JXB2 A0A067QS73 A0A1E1WL78 A0A2P8Y802 D6WFS6 A0A088AJZ1 A0A2A3EK01 A0A154PSY9 A0A310SEA5 A0A0L7RJK7 A0A232ET24 A0A1L8DV56 A0A1L8DVK6 U5EL05 A0A0A9WT81 A0A195CP19 A0A026VZU1 F4WE30 A0A1Q3G2Y3 A0A1Q3G3G6 A0A151K0C2 A0A146LPQ7 Q177Z0 A0A0H3UAL5 A0A3L8D5G8 A0A0L7KI75 Q9VHR6 A0A2M4CRX2 A0A2M4CRU8 B3P4Y2 A0A1J1J630 A0A2M4BKK7 B4PT54 B4HLK7 A0A2M4A1K5 A0A1W4VHP6 A0A2J7PMP5 A0A212FIG9 A0A0M4EES6 B4QYL3 B3M0F1 A0A0M9AC92 A0A0A1X1I5 T1IPJ1 Q298R4 A0A0K8V099 Q7Q6R5 A0A2Y9D0T3 B4LYY6 B4JV55 E0W0V2 A0A151IR64 A0A0L0C0Y2
PDB
5C50
E-value=1.35563e-28,
Score=315
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Under starvation conditions, is localized to puncate structures primarily representing the isolation membrane that sequesters a portion of the cytoplasm resulting in the formation of an autophagosome. With evidence from 1 publications.
Cytosol Under starvation conditions, is localized to puncate structures primarily representing the isolation membrane that sequesters a portion of the cytoplasm resulting in the formation of an autophagosome. With evidence from 1 publications.
Preautophagosomal structure Under starvation conditions, is localized to puncate structures primarily representing the isolation membrane that sequesters a portion of the cytoplasm resulting in the formation of an autophagosome. With evidence from 1 publications.
Cytosol Under starvation conditions, is localized to puncate structures primarily representing the isolation membrane that sequesters a portion of the cytoplasm resulting in the formation of an autophagosome. With evidence from 1 publications.
Preautophagosomal structure Under starvation conditions, is localized to puncate structures primarily representing the isolation membrane that sequesters a portion of the cytoplasm resulting in the formation of an autophagosome. With evidence from 1 publications.
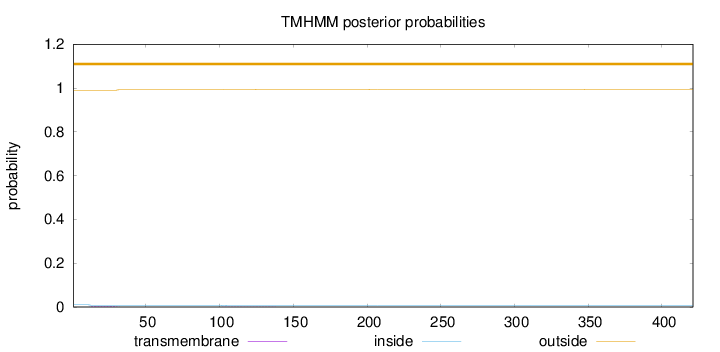
Length:
421
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0607
Exp number, first 60 AAs:
0.04639
Total prob of N-in:
0.01004
outside
1 - 421
Population Genetic Test Statistics
Pi
3.148629
Theta
14.814897
Tajima's D
-1.49968
CLR
0.03653
CSRT
0.0576971151442428
Interpretation
Uncertain
