Gene
KWMTBOMO00516
Pre Gene Modal
BGIBMGA012277
Annotation
Protein_SERAC1_[Operophtera_brumata]
Full name
GPI inositol-deacylase
Location in the cell
Mitochondrial Reliability : 1.67 Nuclear Reliability : 1.218
Sequence
CDS
ATGAGCCTGCAAGACAGACTAAAGCCCATTTTGAAAATACTAAAATTTACATCAATTTGGGGAGGTGGTGGAATTTTTTTTGCTTATCATGTCAGTCAGACGTACAAAACAGTAGACAAGATCGTAAACACAAAAGTTCTAGATCGTGAAAAGAAACATACCCCAGAATATATTTACATTGACGACCCTTCATACAGCATCACATTAAAAAGGGAACAAGACAAAGAATCTAGACGGTCTATAGGTCTCTCGCGAGTTTGGAAGAGTCTGAAGCATTCTTTAGCATGGCGTCTTTTGTGGTTATGTCGTCACGGGAACAAGGAGCAGCGTAACATAGCTCTGAAACAGCTAGCTGCTTTCAAAAACAATAAGATTTGGGACTGTCAAAAACTCGCTCAAGCTTTAGACATGAACACAGCCGTCTTGCTAGCAAGAACTCGCGGTGCTGATCTACGGTACTTCCTCCCTCCGCCTATACATGTACGTAGAGCCGCATTAACTTCTGAATTACTCTCTTTCAAATTCCGAGACATGATACTTTCTGTTCAAGGAGTCCATCCACACAGTTGTATTCAACATTTCCTATCGAAATATTTTGCCAATATTCAGGAACAGGCCTTAGAAGCAGATAATATTCCCGCAAAGCCAGATAGTATTAGTGAGAGAGATTTGTGTATTTTGTGTCTTGATGCCCTCTATCACCACATCAGTCTTTTTCATGTTAACGATTATGAAGATGATGATTCTATAAAAACTCTTATAAAAATGGGACTCCTTCCAAGATTAGCTGAGTTAATGTTAAGAGAGCGCGATGACACTGATTTGGATTTGGCTGTACTTAAAATACTCACTGTTCTGAGTGTTCATTGTAATTTATTATTAGATTTTTTTCAAAACGGTCTTATAAAAGAGTTGTCTCGGCTCCTTAGATCAAAAGATGTGCGATTGGCAAGTTCTGCTGCAGTTTGTTTAGCAAATTTATCAGGGGAACATTGTTACAGACCAGGACTGTACTTGTTGCACCCATTGTATCGAACTACAGTTGCACCAACATGTGACACTCTTCTAGTGCACGGCCTCAGAGGTGGTGTTTTTGTCACATGGCGACAGAGAGATAAAAAGTGCGCTGAACCTCTTGGAATTGTTGAAGGAACAATATCCGATGTTGACTGTGATCCCTGCGAAGAAAAATCAATCGATACAAAGTATCTTGACCCAGATTTGCAACAAGTAATGGATGATCTGATAGAATTAGATGATGAAACTCTGTTGTCTGACTTAGAAGTAGTTTTACATGACCTCCCTATTGAGGCAAAGCGAGAACCTTTGAATACTAATATTTACACTATAAGTAAGAAACGCATAGCCCTTATGCAGGAAGAAGAGGATCAATGTAATTATACATTTTGTTGGCCTAAAGATTGGCTACCTAAAGACTGCAATAATTTGAGAATATTAGGTATCAATTATTGGAGCTCAATATCTGAATGGCTAGAACGATGCCCCCTACAGACAGCTGACATATCAACCAAGGCTTCAGAATTAGGTCCTACATTAGTTGATGCTGGTGTCGGAAGAAAAGGTACTCCTGTTGTGTGGTTAGCTCATTCCATGGGAGGGCTTATAGTTAAACAAGTACTCATTGACTCTGCTCAAAGTAGTGATGACAAATTTAAAAGGCTTTCAAGGAATACTAAGGCTGTATTGTTTTATAGTACACCTCATAAAGGTAGTGCACTTGCGACTATGCCAAGGGCTGCAGCTGCTGTGCTGTGGCCTTCAAACGATGTGCGTCAGTTACAAGAAAACTCGCCAGTTTTACTTGAACTGCATAGAGCATTCATACACGCAGCTGAACAATACCAATGGGAGACCATAAGTTTTGCTGAAATGATGCCAACTTTGGTTACAACATTTAAAGTACCAATCCACTTTGTAGAAGCTTATTCTGCAGACTTAGGACGAGGTGTGTTTTATCAGTTACCTTTAGATCATCTGTCTATATGTAAGCCGGCCACGAGACAATCAATACTGTATACAACAGTCCTGGATGTAATACAAAGGGTGACTGCCAGAAATATTGAAATAAAATACTCTGATTCCCTTATAAAATGGATTATTGACATGCTTTGGTGGACTCTGAGGACAAAAACTAATGAGGCTATAGAAAAAGTTGATGAAGCTCAAAGAATTGAAGGTCTTCGTTGGTATGAGCGAGTCCTACTCGACACATTTACTGATGGATTTACAGATTAG
Protein
MSLQDRLKPILKILKFTSIWGGGGIFFAYHVSQTYKTVDKIVNTKVLDREKKHTPEYIYIDDPSYSITLKREQDKESRRSIGLSRVWKSLKHSLAWRLLWLCRHGNKEQRNIALKQLAAFKNNKIWDCQKLAQALDMNTAVLLARTRGADLRYFLPPPIHVRRAALTSELLSFKFRDMILSVQGVHPHSCIQHFLSKYFANIQEQALEADNIPAKPDSISERDLCILCLDALYHHISLFHVNDYEDDDSIKTLIKMGLLPRLAELMLRERDDTDLDLAVLKILTVLSVHCNLLLDFFQNGLIKELSRLLRSKDVRLASSAAVCLANLSGEHCYRPGLYLLHPLYRTTVAPTCDTLLVHGLRGGVFVTWRQRDKKCAEPLGIVEGTISDVDCDPCEEKSIDTKYLDPDLQQVMDDLIELDDETLLSDLEVVLHDLPIEAKREPLNTNIYTISKKRIALMQEEEDQCNYTFCWPKDWLPKDCNNLRILGINYWSSISEWLERCPLQTADISTKASELGPTLVDAGVGRKGTPVVWLAHSMGGLIVKQVLIDSAQSSDDKFKRLSRNTKAVLFYSTPHKGSALATMPRAAAAVLWPSNDVRQLQENSPVLLELHRAFIHAAEQYQWETISFAEMMPTLVTTFKVPIHFVEAYSADLGRGVFYQLPLDHLSICKPATRQSILYTTVLDVIQRVTARNIEIKYSDSLIKWIIDMLWWTLRTKTNEAIEKVDEAQRIEGLRWYERVLLDTFTDGFTD
Summary
Description
Involved in inositol deacylation of GPI-anchored proteins which plays important roles in the quality control and ER-associated degradation of GPI-anchored proteins.
Similarity
Belongs to the GPI inositol-deacylase family.
Uniprot
H9JRW7
A0A0L7L078
A0A2A4JE37
A0A2H1VCF9
A0A194QU20
A0A194PL50
+ More
A0A212EIE5 A0A2J7Q0Y4 A0A2J7Q0X1 A0A067QU96 K7IRT0 A0A232FF61 A0A158ND75 A0A2A3ENP0 A0A151WG90 E9J2D1 A0A087ZZG8 A0A0L7QNP2 A0A195B1B6 A0A2J7Q0Y8 A0A0J7NRW5 A0A195DKA7 A0A151I8R0 A0A195FHN0 A0A2M4CPW0 A0A2M4A0Q7 A0A154PQ71 E2A031 F4X787 A0A2M3ZG89 A0A3L8D3K6 A0A310SPK4 A0A1Y1LZ43 A0A182GCC9 A0A1Q3FEG5 A0A139WN90 A0A0M9A4Y0 B0W7Z2 A0A026WXE3 U5EDY0 A0A2M4A0Y5 T1E1U1 A0A2M4A0N7 A0A2P8XKB0 U4U596 A0A2R7W0M9 A0A2J7Q0X2 A0A2H8TF92 J9JL18 E0VG14 A0A2S2QD03 B3MM44 T1H7Y1 A0A2J7Q0V6 A0A2J7Q0Y0 N6SXA2
A0A212EIE5 A0A2J7Q0Y4 A0A2J7Q0X1 A0A067QU96 K7IRT0 A0A232FF61 A0A158ND75 A0A2A3ENP0 A0A151WG90 E9J2D1 A0A087ZZG8 A0A0L7QNP2 A0A195B1B6 A0A2J7Q0Y8 A0A0J7NRW5 A0A195DKA7 A0A151I8R0 A0A195FHN0 A0A2M4CPW0 A0A2M4A0Q7 A0A154PQ71 E2A031 F4X787 A0A2M3ZG89 A0A3L8D3K6 A0A310SPK4 A0A1Y1LZ43 A0A182GCC9 A0A1Q3FEG5 A0A139WN90 A0A0M9A4Y0 B0W7Z2 A0A026WXE3 U5EDY0 A0A2M4A0Y5 T1E1U1 A0A2M4A0N7 A0A2P8XKB0 U4U596 A0A2R7W0M9 A0A2J7Q0X2 A0A2H8TF92 J9JL18 E0VG14 A0A2S2QD03 B3MM44 T1H7Y1 A0A2J7Q0V6 A0A2J7Q0Y0 N6SXA2
EC Number
3.1.-.-
Pubmed
EMBL
BABH01017024
JTDY01004017
KOB68706.1
NWSH01001921
PCG69672.1
ODYU01001792
+ More
SOQ38476.1 KQ461108 KPJ09028.1 KQ459605 KPI91830.1 AGBW02014676 OWR41240.1 NEVH01019960 PNF22235.1 PNF22233.1 KK852945 KDR13447.1 NNAY01000331 OXU29160.1 ADTU01012470 ADTU01012471 KZ288212 PBC32816.1 KQ983185 KYQ46852.1 GL767845 EFZ13006.1 KQ414855 KOC60106.1 KQ976690 KYM78080.1 PNF22236.1 LBMM01002178 KMQ95175.1 KQ980800 KYN12914.1 KQ978338 KYM95025.1 KQ981560 KYN39893.1 GGFL01003169 MBW67347.1 GGFK01001010 MBW34331.1 KQ435034 KZC14051.1 GL435543 EFN73214.1 GL888828 EGI57699.1 GGFM01006752 MBW27503.1 QOIP01000014 RLU14829.1 KQ761906 OAD56460.1 GEZM01048952 JAV76247.1 JXUM01009077 KQ560275 KXJ83384.1 GFDL01009095 JAV25950.1 KQ971312 KYB29402.1 KQ435732 KOX77317.1 DS231856 EDS38454.1 KK107070 EZA60692.1 GANO01004548 JAB55323.1 GGFK01000977 MBW34298.1 GALA01001765 JAA93087.1 GGFK01001055 MBW34376.1 PYGN01001864 PSN32433.1 KB631665 ERL85140.1 KK854169 PTY12490.1 PNF22231.1 GFXV01000905 MBW12710.1 ABLF02034499 DS235131 EEB12320.1 GGMS01006421 MBY75624.1 CH902620 EDV30859.1 ACPB03025021 PNF22232.1 PNF22234.1 APGK01053205 KB741222 ENN72409.1
SOQ38476.1 KQ461108 KPJ09028.1 KQ459605 KPI91830.1 AGBW02014676 OWR41240.1 NEVH01019960 PNF22235.1 PNF22233.1 KK852945 KDR13447.1 NNAY01000331 OXU29160.1 ADTU01012470 ADTU01012471 KZ288212 PBC32816.1 KQ983185 KYQ46852.1 GL767845 EFZ13006.1 KQ414855 KOC60106.1 KQ976690 KYM78080.1 PNF22236.1 LBMM01002178 KMQ95175.1 KQ980800 KYN12914.1 KQ978338 KYM95025.1 KQ981560 KYN39893.1 GGFL01003169 MBW67347.1 GGFK01001010 MBW34331.1 KQ435034 KZC14051.1 GL435543 EFN73214.1 GL888828 EGI57699.1 GGFM01006752 MBW27503.1 QOIP01000014 RLU14829.1 KQ761906 OAD56460.1 GEZM01048952 JAV76247.1 JXUM01009077 KQ560275 KXJ83384.1 GFDL01009095 JAV25950.1 KQ971312 KYB29402.1 KQ435732 KOX77317.1 DS231856 EDS38454.1 KK107070 EZA60692.1 GANO01004548 JAB55323.1 GGFK01000977 MBW34298.1 GALA01001765 JAA93087.1 GGFK01001055 MBW34376.1 PYGN01001864 PSN32433.1 KB631665 ERL85140.1 KK854169 PTY12490.1 PNF22231.1 GFXV01000905 MBW12710.1 ABLF02034499 DS235131 EEB12320.1 GGMS01006421 MBY75624.1 CH902620 EDV30859.1 ACPB03025021 PNF22232.1 PNF22234.1 APGK01053205 KB741222 ENN72409.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000053240
UP000053268
UP000007151
+ More
UP000235965 UP000027135 UP000002358 UP000215335 UP000005205 UP000242457 UP000075809 UP000005203 UP000053825 UP000078540 UP000036403 UP000078492 UP000078542 UP000078541 UP000076502 UP000000311 UP000007755 UP000279307 UP000069940 UP000249989 UP000007266 UP000053105 UP000002320 UP000053097 UP000245037 UP000030742 UP000007819 UP000009046 UP000007801 UP000015103 UP000019118
UP000235965 UP000027135 UP000002358 UP000215335 UP000005205 UP000242457 UP000075809 UP000005203 UP000053825 UP000078540 UP000036403 UP000078492 UP000078542 UP000078541 UP000076502 UP000000311 UP000007755 UP000279307 UP000069940 UP000249989 UP000007266 UP000053105 UP000002320 UP000053097 UP000245037 UP000030742 UP000007819 UP000009046 UP000007801 UP000015103 UP000019118
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JRW7
A0A0L7L078
A0A2A4JE37
A0A2H1VCF9
A0A194QU20
A0A194PL50
+ More
A0A212EIE5 A0A2J7Q0Y4 A0A2J7Q0X1 A0A067QU96 K7IRT0 A0A232FF61 A0A158ND75 A0A2A3ENP0 A0A151WG90 E9J2D1 A0A087ZZG8 A0A0L7QNP2 A0A195B1B6 A0A2J7Q0Y8 A0A0J7NRW5 A0A195DKA7 A0A151I8R0 A0A195FHN0 A0A2M4CPW0 A0A2M4A0Q7 A0A154PQ71 E2A031 F4X787 A0A2M3ZG89 A0A3L8D3K6 A0A310SPK4 A0A1Y1LZ43 A0A182GCC9 A0A1Q3FEG5 A0A139WN90 A0A0M9A4Y0 B0W7Z2 A0A026WXE3 U5EDY0 A0A2M4A0Y5 T1E1U1 A0A2M4A0N7 A0A2P8XKB0 U4U596 A0A2R7W0M9 A0A2J7Q0X2 A0A2H8TF92 J9JL18 E0VG14 A0A2S2QD03 B3MM44 T1H7Y1 A0A2J7Q0V6 A0A2J7Q0Y0 N6SXA2
A0A212EIE5 A0A2J7Q0Y4 A0A2J7Q0X1 A0A067QU96 K7IRT0 A0A232FF61 A0A158ND75 A0A2A3ENP0 A0A151WG90 E9J2D1 A0A087ZZG8 A0A0L7QNP2 A0A195B1B6 A0A2J7Q0Y8 A0A0J7NRW5 A0A195DKA7 A0A151I8R0 A0A195FHN0 A0A2M4CPW0 A0A2M4A0Q7 A0A154PQ71 E2A031 F4X787 A0A2M3ZG89 A0A3L8D3K6 A0A310SPK4 A0A1Y1LZ43 A0A182GCC9 A0A1Q3FEG5 A0A139WN90 A0A0M9A4Y0 B0W7Z2 A0A026WXE3 U5EDY0 A0A2M4A0Y5 T1E1U1 A0A2M4A0N7 A0A2P8XKB0 U4U596 A0A2R7W0M9 A0A2J7Q0X2 A0A2H8TF92 J9JL18 E0VG14 A0A2S2QD03 B3MM44 T1H7Y1 A0A2J7Q0V6 A0A2J7Q0Y0 N6SXA2
Ontologies
GO
PANTHER
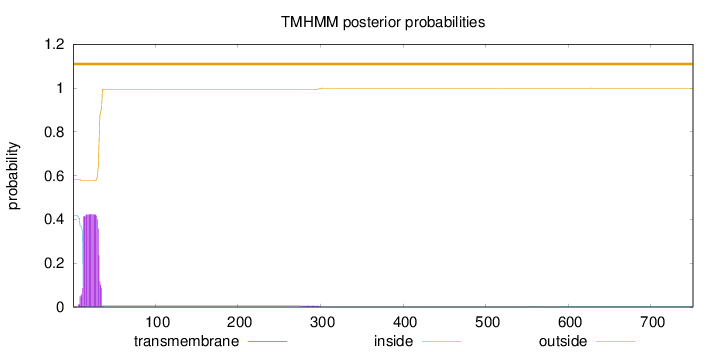
Topology
Subcellular location
Endoplasmic reticulum membrane
Length:
751
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
8.85017999999996
Exp number, first 60 AAs:
8.71722
Total prob of N-in:
0.41764
outside
1 - 751
Population Genetic Test Statistics
Pi
0.444541
Theta
1.711649
Tajima's D
-1.649947
CLR
0.000511
CSRT
0.031598420078996
Interpretation
Possibly Positive selection
