Gene
KWMTBOMO00514
Pre Gene Modal
BGIBMGA012318
Annotation
PREDICTED:_tyrosine-protein_kinase_PR2_[Papilio_machaon]
Location in the cell
Nuclear Reliability : 4.142
Sequence
CDS
ATGGAAAGTATAAGCGACGATCGAGAGAACTTAAAGGATTTTCTTGAAGAAGCTGAACTTCAACAATACCATGAGCTCTTTAGAGATGTCTTGAAAGTGACAAAACTATCGCAATTAAAATTCGTCGTGACAGAGGACTTGGTACAAATTGGCTTCTCGAAACCAGAACAAAGACGATATAAAAAAATATATTCCAAATATTTTCCAAATCCATACATCACCAAAATAAAGAGACTGCTTAGCTCTAGTAAAAAGAATGATCCGTTTTCACATGGACAATCGACGATACCTGTCGATTTGCAACCGTTTTCAGACAGCACCGTTAAAATTCCCACAAAACATATAATATCAATAGAAGATATAACTATTAATAAAGAGCTTGGCATGGGCCAGTTTGGTGTTGTGCAACAAGGCACATGGACTACAGGGAATCAAAGACTGCAAGTTGCAATAAAATGTTTGGGAAACGAACGTATGACATCAAATTCAACTGAGTTCCTAAAGGAGGCAGCCGTGATGCACAGCATAGAGCATCCAAATATAGTGCGATTGTTTGGCGTCGTTCTGCATTTAGATTCCTTGATGTTGGTAACAGAGCTGGCTCCTCTCCGTTCACTGCTGGAGTGCCTCCGTGAAGTGTCACTCCGGCCCAATTTCCCCGTTCCTACTCTCTGCGAGTTCGCCGAACAGATCTGTGATGGCATGACTTATTTAGAAAACAAAAGACTGATTCATAGGGATTTAGCAGCAAGAAATATTTTAGTATTCAACAAAGATAGAGTAAAGATTTCAGACTTTGGTCTGTCGCGCGCACTTGGTGTCGGCAAGGATTACTATCAGACGAATTATAATGTCAATCTTAAATTGCCGGTGGCTTGGTGCGCGCCAGAATGTATATTGTACTTACGTTTTACGTCATCGAGCGACGTCTGGGCGTTCGGTGTTTGTCTTTGGGAGATGTTCACATATGGCTTCCAACCGTGGGCGGCTTTCTCCGGACAGCAGATATTAGAAGCTATTGATACGCCTAATTTTCAGAGATTGGAAAGACCGGAGTGCTGTCCCGAAGCGTATTATTCGACGATGCTGGAGTGCTGGTCGCACGAGCCAAACGATCGGCCGAAATTCAAGGACCTGGGACCAAAGTTACTCGGTATACGACCTGAAATGGTTCAAGCAACATCGAACTTCGTGAGAACAGACGACGGAAGACGCACTAATTATTTAGATTATAAAGTTGGACAGATCATTACCGTTTTAGGAAAAGAAGGCATGACTAAGAGCTCCTTGTGGTACGGAGTTCTCCCCGGCGGTGCCTGCGGTGTGTTCGATCCTAACCAGACCAAACCATATTCGAAACCAGAGAAGCCATTATCGCTGGGCTCGCTATCGCCGAATCCGAATCGTCATTCGTCACGTTCGTTACGTCCCTCCCTACTGCGCTCGGACGCGAAGCGACACACGTACACCGGGAAACGGACGATACAACGCAACATGATATCTAGTCCGCAAGGCGACGTCAAACACACGGGACACGTAGGACTCGACGGCGCATATTTCGGAGACATATCGTTCTTGGACCCGGCCGGTTGCCCGCCCAGACAAGTGGTGACTCCGTACAAGCCCTCGGAGGACTTGGAGCAGGTTCCCCTGATCAACCCGAACTCCCCTTCACACCTGACCGGGGGAGCCTCGAGTGCGCCTCCGTATCCGTTGCTGTCGTCACGTCCGGATCACAGGAACGATTGCGATGAAGTCGACCTGGCTTATGGCGAAAGTCGAAGTAAAGCTCAGAAGAAAAACAATCACAAGAATTCGGAAACCACCGCCTGTAAAAATATATTGAATAAGGTGAAAAGCGCGACTATTCGTCGTTCAAATAAAACAGTGGACGCAAACAATAAAACGGACGAGGTTCACGAATACCATGAAATATCCGACACTGATATGGAATGTCCACATCCTGCGAGTGCGAGGAATGAACATCTTGGATCACCCGAAAGTCCAGGGATTATTAAAGGCTATACAGACTTCAGTCAGAGCCTCATCGATGAGATGGAAACTATGTTCCGAAATCTGGACGCTAAGAGACGTGACGAGCCGGGCACATCGAGTAAGCATTTCGACATCGATGCATTGAGATCTCATACATTGTCGAAGTTTGATAAGAAGAAGAACTTCAACACCGGCACAATGAAACCAATGTCGGCCCACGATGAGAAGACACTGGAAACGGCGATGTCCATGGCCAATGAACTGACCACAAAGTCAATGACCGATCTGGGCGGTGACAACAAGTTGCCAACATCGCCAAACGATAAATCCAAGTTTCACTTCAAATTCCCGCAGCTGTCTATACATCACGAGAAACCTAATGAAGTGATTGACAAAGATGCTTTGGCTAAACAAGAACGTCGCAACTTCAGCGAGGAAGCTAAAAGCGTACCCGACATACAGTCGACAATAACGAAAGAGAGCAAAGCTGCATACACCCATTTAATCGAGGAGCCGACGCCCTCTGGATCATTGTTGACTCAAATCGCCAAAGAAACGAAAGCGAAGGGTCATCACCAGCCGGCAACAATGCCCAAACCAGCACCGAGAAGCGAACCGCGTCCTAGATTCCATTCACTGCAGATCGCACCACCATGCAAACCTAGGCCGTGTCCTGAAGTCGAAACAATTGAAAAAGAAATCACAAATCCATTACCGCTACCACCGAGAGCTACCAAACCAAAACTGGTCGACAAACGACGTCACATCCGAAAGTATCCCCTCAAGCTGCCATCGGACATGGTTCTCCCCGAGCCCGGTCCAAGCAAGCAGATAATATACCAAAACGCTTTCACTCCGGTTAACTGTCGCTTGCAACACGACAACGATCGCAAGAGTAACGGCGTCGACGTAATTGACGGTCCAGCTAAAGGAACGAATCCGTTCTCTCGTTCCGGACCTAGTTCGGTTAATCCCTTCGATAATTGTATCGAAACGAACGACGACGGCATCGATCTGCATCTCGACGTTGTCCGGTACGACGTTAGTGAAGACGAAGACGACGAACATGACATCGCCGAAGGAGCTCTTGTTAAAGAATGTTCTGACCACGTGTCGGTCGAGGATCTGCTCGAGTTCGCCGACCAGAAGCCCTGCGGCAGACAACGTGGCGTCGAATCCGATGAAGTCAGGATCATGAGAAAAGTACTTAATCAGAAAGTGACACCTGAAGAATGCTTAGCCGCTTTAGAGCTGAGTGATTGGAACGTTCACCAAGCCATCAAAGTGATGCGCGTCAAGATATCTGTCGGCGAGGCTGTTTCCCTGGAGGAATGTAGCACGGAGCTGCTAAAAGCCTGCGGAGATATTGTTAAGGCTTCGGCCGTACTAGTCAGGTTTGGTTTTAGGAATGCTGATAGCGAAAAATTGTTGAAGAAGTTACAATGA
Protein
MESISDDRENLKDFLEEAELQQYHELFRDVLKVTKLSQLKFVVTEDLVQIGFSKPEQRRYKKIYSKYFPNPYITKIKRLLSSSKKNDPFSHGQSTIPVDLQPFSDSTVKIPTKHIISIEDITINKELGMGQFGVVQQGTWTTGNQRLQVAIKCLGNERMTSNSTEFLKEAAVMHSIEHPNIVRLFGVVLHLDSLMLVTELAPLRSLLECLREVSLRPNFPVPTLCEFAEQICDGMTYLENKRLIHRDLAARNILVFNKDRVKISDFGLSRALGVGKDYYQTNYNVNLKLPVAWCAPECILYLRFTSSSDVWAFGVCLWEMFTYGFQPWAAFSGQQILEAIDTPNFQRLERPECCPEAYYSTMLECWSHEPNDRPKFKDLGPKLLGIRPEMVQATSNFVRTDDGRRTNYLDYKVGQIITVLGKEGMTKSSLWYGVLPGGACGVFDPNQTKPYSKPEKPLSLGSLSPNPNRHSSRSLRPSLLRSDAKRHTYTGKRTIQRNMISSPQGDVKHTGHVGLDGAYFGDISFLDPAGCPPRQVVTPYKPSEDLEQVPLINPNSPSHLTGGASSAPPYPLLSSRPDHRNDCDEVDLAYGESRSKAQKKNNHKNSETTACKNILNKVKSATIRRSNKTVDANNKTDEVHEYHEISDTDMECPHPASARNEHLGSPESPGIIKGYTDFSQSLIDEMETMFRNLDAKRRDEPGTSSKHFDIDALRSHTLSKFDKKKNFNTGTMKPMSAHDEKTLETAMSMANELTTKSMTDLGGDNKLPTSPNDKSKFHFKFPQLSIHHEKPNEVIDKDALAKQERRNFSEEAKSVPDIQSTITKESKAAYTHLIEEPTPSGSLLTQIAKETKAKGHHQPATMPKPAPRSEPRPRFHSLQIAPPCKPRPCPEVETIEKEITNPLPLPPRATKPKLVDKRRHIRKYPLKLPSDMVLPEPGPSKQIIYQNAFTPVNCRLQHDNDRKSNGVDVIDGPAKGTNPFSRSGPSSVNPFDNCIETNDDGIDLHLDVVRYDVSEDEDDEHDIAEGALVKECSDHVSVEDLLEFADQKPCGRQRGVESDEVRIMRKVLNQKVTPEECLAALELSDWNVHQAIKVMRVKISVGEAVSLEECSTELLKACGDIVKASAVLVRFGFRNADSEKLLKKLQ
Summary
Similarity
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Uniprot
A0A2A4K4X2
A0A194QU81
A0A194PEX5
A0A212EIC9
A0A1B6DZ52
A0A147BPH3
+ More
A0A0P5CBR9 A0A0P5BZT6 A0A164YHP4 A0A0P5ZYQ2 A0A0P5XGN2 A0A0P4ZWW3 A0A0P5E3S5 A0A0P5ZR24 A0A2R5LKJ1 A0A0N8EB83 A0A0P5WFF6 A0A0P5SNZ8 A0A0P5FEE7 A0A0N7ZU53 A0A0P5DPJ2 A0A0P5EL36 J9K951 A0A0P5CZ27 A0A0P6E2I8 A0A131Z2J1 A0A224YWW9 L7M1F3 E9FQQ5 A0A0P5C0Z9 A0A293LPV6 A0A0P5C0Y6 A0A0P6AZ62 A0A0P5DZX0 A0A1E1XPT2 A0A131XFP4 A0A2H8TXI1 K7IMP9 A0A158NJI7 Q16R80 A0A0C9QTQ7 A0A0C9RB13 A0A1L8DKJ7 A0A067R0F6 A0A2S2PCZ7 A0A1S4FS22 E0VHV9 A0A026X2E1 A0A2J7QAL9 A0A2A3EQH2 A0A087U5M2 B4J4Z7 A0A3B0JM64 A0A087ZNU3 A0A3Q0J314 B4HQ06 A0A0J9RCN6 A0A0Q5VN54 B3NRR4 Q7PT15 T1ISC4 A0A1W4VQE7 A0A1B6DJ21 A0A0P5TID5 A0A0P5BB28 A0A0P5AM61 A0A0P5BHA9 B4MPB9 A0A0P5ZNN8 B4LMI5 B4P4U8 B4KSW8 A0A0P5BZN1 A0A1E1X525 A0A0P5ZRJ7 A0A1Q3G110 A0A0P5UF42 A0A0P4VZY0 A0A0P4VUU8 A0A1Q3G109 A0A0N7ZUH2 A0A0P5Y1A6 A0A0P5Z2M3 A0A0P4WEW9 A0A182IUJ2 A0A182QNF6 A0A034V5D9 A0A182KS24 A0A0K8U9Y3 A0A0A1WHQ6 A0A1W4VCC3 A0A0K8UR12 A0A182M8B8 A0A2M4CR01 W5JKS0 A0A2M4CQX7 A0A182RB12
A0A0P5CBR9 A0A0P5BZT6 A0A164YHP4 A0A0P5ZYQ2 A0A0P5XGN2 A0A0P4ZWW3 A0A0P5E3S5 A0A0P5ZR24 A0A2R5LKJ1 A0A0N8EB83 A0A0P5WFF6 A0A0P5SNZ8 A0A0P5FEE7 A0A0N7ZU53 A0A0P5DPJ2 A0A0P5EL36 J9K951 A0A0P5CZ27 A0A0P6E2I8 A0A131Z2J1 A0A224YWW9 L7M1F3 E9FQQ5 A0A0P5C0Z9 A0A293LPV6 A0A0P5C0Y6 A0A0P6AZ62 A0A0P5DZX0 A0A1E1XPT2 A0A131XFP4 A0A2H8TXI1 K7IMP9 A0A158NJI7 Q16R80 A0A0C9QTQ7 A0A0C9RB13 A0A1L8DKJ7 A0A067R0F6 A0A2S2PCZ7 A0A1S4FS22 E0VHV9 A0A026X2E1 A0A2J7QAL9 A0A2A3EQH2 A0A087U5M2 B4J4Z7 A0A3B0JM64 A0A087ZNU3 A0A3Q0J314 B4HQ06 A0A0J9RCN6 A0A0Q5VN54 B3NRR4 Q7PT15 T1ISC4 A0A1W4VQE7 A0A1B6DJ21 A0A0P5TID5 A0A0P5BB28 A0A0P5AM61 A0A0P5BHA9 B4MPB9 A0A0P5ZNN8 B4LMI5 B4P4U8 B4KSW8 A0A0P5BZN1 A0A1E1X525 A0A0P5ZRJ7 A0A1Q3G110 A0A0P5UF42 A0A0P4VZY0 A0A0P4VUU8 A0A1Q3G109 A0A0N7ZUH2 A0A0P5Y1A6 A0A0P5Z2M3 A0A0P4WEW9 A0A182IUJ2 A0A182QNF6 A0A034V5D9 A0A182KS24 A0A0K8U9Y3 A0A0A1WHQ6 A0A1W4VCC3 A0A0K8UR12 A0A182M8B8 A0A2M4CR01 W5JKS0 A0A2M4CQX7 A0A182RB12
Pubmed
EMBL
NWSH01000146
PCG79076.1
KQ461108
KPJ09027.1
KQ459605
KPI91832.1
+ More
AGBW02014676 OWR41239.1 GEDC01009362 GEDC01006353 JAS27936.1 JAS30945.1 GEGO01002726 JAR92678.1 GDIP01177683 JAJ45719.1 GDIP01177682 JAJ45720.1 LRGB01000915 KZS15259.1 GDIP01037892 JAM65823.1 GDIP01094130 GDIP01072380 JAM31335.1 GDIP01210259 GDIP01179613 GDIP01147475 GDIP01131821 GDIP01052145 JAJ13143.1 GDIP01147474 GDIP01133322 GDIP01131822 GDIP01052146 JAJ75928.1 GDIP01041235 JAM62480.1 GGLE01005906 MBY10032.1 GDIQ01043804 JAN50933.1 GDIP01137323 GDIP01090074 GDIP01088132 GDIP01087130 JAM16585.1 GDIP01139673 GDIP01137324 GDIP01122473 JAL66390.1 GDIP01212314 GDIP01184573 GDIP01148823 GDIP01044242 JAJ74579.1 GDIP01212313 GDIP01184574 GDIP01148822 GDIP01085973 JAJ11089.1 GDIP01153172 JAJ70230.1 GDIP01158222 JAJ65180.1 ABLF02017311 ABLF02017313 GDIP01163474 JAJ59928.1 GDIQ01068743 JAN25994.1 GEDV01003389 JAP85168.1 GFPF01010972 MAA22118.1 GACK01006903 JAA58131.1 GL732523 EFX90330.1 GDIP01177684 JAJ45718.1 GFWV01010788 MAA35517.1 GDIP01177685 JAJ45717.1 GDIP01022360 JAM81355.1 GDIP01154670 JAJ68732.1 GFAA01002358 JAU01077.1 GEFH01004115 JAP64466.1 GFXV01006805 MBW18610.1 ADTU01017908 ADTU01017909 ADTU01017910 ADTU01017911 ADTU01017912 ADTU01017913 ADTU01017914 ADTU01017915 ADTU01017916 ADTU01017917 CH477722 EAT36899.1 GBYB01004062 JAG73829.1 GBYB01004061 JAG73828.1 GFDF01007095 JAV06989.1 KK852804 KDR16216.1 GGMR01014712 MBY27331.1 DS235172 EEB12965.1 KK107020 EZA62457.1 NEVH01016330 PNF25622.1 KZ288204 PBC33291.1 KK118303 KFM72661.1 CH916367 EDW01703.1 OUUW01000001 SPP74366.1 CH480816 EDW47669.1 CM002911 KMY93399.1 CH954179 KQS62774.1 EDV56216.2 AAAB01008807 EAA04689.5 AFFK01018942 GEDC01011680 JAS25618.1 GDIP01125826 JAL77888.1 GDIP01186970 JAJ36432.1 GDIP01201869 JAJ21533.1 GDIP01184575 JAJ38827.1 CH963849 EDW73958.1 GDIP01041236 JAM62479.1 CH940648 EDW59972.2 CM000158 EDW90669.2 CH933808 EDW08465.2 GDIP01177736 JAJ45666.1 GFAC01004818 JAT94370.1 GDIP01039290 JAM64425.1 GFDL01001556 JAV33489.1 GDIP01113932 JAL89782.1 GDRN01095215 JAI59585.1 GDRN01095214 JAI59586.1 GFDL01001560 JAV33485.1 GDIP01211361 JAJ12041.1 GDIP01065379 JAM38336.1 GDIP01050890 JAM52825.1 GDRN01095210 JAI59588.1 AXCN02000894 GAKP01022204 JAC36748.1 GDHF01028837 JAI23477.1 GBXI01015708 JAC98583.1 GDHF01023361 JAI28953.1 AXCM01000589 GGFL01003555 MBW67733.1 ADMH02001258 ETN63369.1 GGFL01003556 MBW67734.1
AGBW02014676 OWR41239.1 GEDC01009362 GEDC01006353 JAS27936.1 JAS30945.1 GEGO01002726 JAR92678.1 GDIP01177683 JAJ45719.1 GDIP01177682 JAJ45720.1 LRGB01000915 KZS15259.1 GDIP01037892 JAM65823.1 GDIP01094130 GDIP01072380 JAM31335.1 GDIP01210259 GDIP01179613 GDIP01147475 GDIP01131821 GDIP01052145 JAJ13143.1 GDIP01147474 GDIP01133322 GDIP01131822 GDIP01052146 JAJ75928.1 GDIP01041235 JAM62480.1 GGLE01005906 MBY10032.1 GDIQ01043804 JAN50933.1 GDIP01137323 GDIP01090074 GDIP01088132 GDIP01087130 JAM16585.1 GDIP01139673 GDIP01137324 GDIP01122473 JAL66390.1 GDIP01212314 GDIP01184573 GDIP01148823 GDIP01044242 JAJ74579.1 GDIP01212313 GDIP01184574 GDIP01148822 GDIP01085973 JAJ11089.1 GDIP01153172 JAJ70230.1 GDIP01158222 JAJ65180.1 ABLF02017311 ABLF02017313 GDIP01163474 JAJ59928.1 GDIQ01068743 JAN25994.1 GEDV01003389 JAP85168.1 GFPF01010972 MAA22118.1 GACK01006903 JAA58131.1 GL732523 EFX90330.1 GDIP01177684 JAJ45718.1 GFWV01010788 MAA35517.1 GDIP01177685 JAJ45717.1 GDIP01022360 JAM81355.1 GDIP01154670 JAJ68732.1 GFAA01002358 JAU01077.1 GEFH01004115 JAP64466.1 GFXV01006805 MBW18610.1 ADTU01017908 ADTU01017909 ADTU01017910 ADTU01017911 ADTU01017912 ADTU01017913 ADTU01017914 ADTU01017915 ADTU01017916 ADTU01017917 CH477722 EAT36899.1 GBYB01004062 JAG73829.1 GBYB01004061 JAG73828.1 GFDF01007095 JAV06989.1 KK852804 KDR16216.1 GGMR01014712 MBY27331.1 DS235172 EEB12965.1 KK107020 EZA62457.1 NEVH01016330 PNF25622.1 KZ288204 PBC33291.1 KK118303 KFM72661.1 CH916367 EDW01703.1 OUUW01000001 SPP74366.1 CH480816 EDW47669.1 CM002911 KMY93399.1 CH954179 KQS62774.1 EDV56216.2 AAAB01008807 EAA04689.5 AFFK01018942 GEDC01011680 JAS25618.1 GDIP01125826 JAL77888.1 GDIP01186970 JAJ36432.1 GDIP01201869 JAJ21533.1 GDIP01184575 JAJ38827.1 CH963849 EDW73958.1 GDIP01041236 JAM62479.1 CH940648 EDW59972.2 CM000158 EDW90669.2 CH933808 EDW08465.2 GDIP01177736 JAJ45666.1 GFAC01004818 JAT94370.1 GDIP01039290 JAM64425.1 GFDL01001556 JAV33489.1 GDIP01113932 JAL89782.1 GDRN01095215 JAI59585.1 GDRN01095214 JAI59586.1 GFDL01001560 JAV33485.1 GDIP01211361 JAJ12041.1 GDIP01065379 JAM38336.1 GDIP01050890 JAM52825.1 GDRN01095210 JAI59588.1 AXCN02000894 GAKP01022204 JAC36748.1 GDHF01028837 JAI23477.1 GBXI01015708 JAC98583.1 GDHF01023361 JAI28953.1 AXCM01000589 GGFL01003555 MBW67733.1 ADMH02001258 ETN63369.1 GGFL01003556 MBW67734.1
Proteomes
UP000218220
UP000053240
UP000053268
UP000007151
UP000076858
UP000007819
+ More
UP000000305 UP000002358 UP000005205 UP000008820 UP000027135 UP000009046 UP000053097 UP000235965 UP000242457 UP000054359 UP000001070 UP000268350 UP000005203 UP000079169 UP000001292 UP000008711 UP000007062 UP000192221 UP000007798 UP000008792 UP000002282 UP000009192 UP000075880 UP000075886 UP000075882 UP000075883 UP000000673 UP000075900
UP000000305 UP000002358 UP000005205 UP000008820 UP000027135 UP000009046 UP000053097 UP000235965 UP000242457 UP000054359 UP000001070 UP000268350 UP000005203 UP000079169 UP000001292 UP000008711 UP000007062 UP000192221 UP000007798 UP000008792 UP000002282 UP000009192 UP000075880 UP000075886 UP000075882 UP000075883 UP000000673 UP000075900
Interpro
CDD
ProteinModelPortal
A0A2A4K4X2
A0A194QU81
A0A194PEX5
A0A212EIC9
A0A1B6DZ52
A0A147BPH3
+ More
A0A0P5CBR9 A0A0P5BZT6 A0A164YHP4 A0A0P5ZYQ2 A0A0P5XGN2 A0A0P4ZWW3 A0A0P5E3S5 A0A0P5ZR24 A0A2R5LKJ1 A0A0N8EB83 A0A0P5WFF6 A0A0P5SNZ8 A0A0P5FEE7 A0A0N7ZU53 A0A0P5DPJ2 A0A0P5EL36 J9K951 A0A0P5CZ27 A0A0P6E2I8 A0A131Z2J1 A0A224YWW9 L7M1F3 E9FQQ5 A0A0P5C0Z9 A0A293LPV6 A0A0P5C0Y6 A0A0P6AZ62 A0A0P5DZX0 A0A1E1XPT2 A0A131XFP4 A0A2H8TXI1 K7IMP9 A0A158NJI7 Q16R80 A0A0C9QTQ7 A0A0C9RB13 A0A1L8DKJ7 A0A067R0F6 A0A2S2PCZ7 A0A1S4FS22 E0VHV9 A0A026X2E1 A0A2J7QAL9 A0A2A3EQH2 A0A087U5M2 B4J4Z7 A0A3B0JM64 A0A087ZNU3 A0A3Q0J314 B4HQ06 A0A0J9RCN6 A0A0Q5VN54 B3NRR4 Q7PT15 T1ISC4 A0A1W4VQE7 A0A1B6DJ21 A0A0P5TID5 A0A0P5BB28 A0A0P5AM61 A0A0P5BHA9 B4MPB9 A0A0P5ZNN8 B4LMI5 B4P4U8 B4KSW8 A0A0P5BZN1 A0A1E1X525 A0A0P5ZRJ7 A0A1Q3G110 A0A0P5UF42 A0A0P4VZY0 A0A0P4VUU8 A0A1Q3G109 A0A0N7ZUH2 A0A0P5Y1A6 A0A0P5Z2M3 A0A0P4WEW9 A0A182IUJ2 A0A182QNF6 A0A034V5D9 A0A182KS24 A0A0K8U9Y3 A0A0A1WHQ6 A0A1W4VCC3 A0A0K8UR12 A0A182M8B8 A0A2M4CR01 W5JKS0 A0A2M4CQX7 A0A182RB12
A0A0P5CBR9 A0A0P5BZT6 A0A164YHP4 A0A0P5ZYQ2 A0A0P5XGN2 A0A0P4ZWW3 A0A0P5E3S5 A0A0P5ZR24 A0A2R5LKJ1 A0A0N8EB83 A0A0P5WFF6 A0A0P5SNZ8 A0A0P5FEE7 A0A0N7ZU53 A0A0P5DPJ2 A0A0P5EL36 J9K951 A0A0P5CZ27 A0A0P6E2I8 A0A131Z2J1 A0A224YWW9 L7M1F3 E9FQQ5 A0A0P5C0Z9 A0A293LPV6 A0A0P5C0Y6 A0A0P6AZ62 A0A0P5DZX0 A0A1E1XPT2 A0A131XFP4 A0A2H8TXI1 K7IMP9 A0A158NJI7 Q16R80 A0A0C9QTQ7 A0A0C9RB13 A0A1L8DKJ7 A0A067R0F6 A0A2S2PCZ7 A0A1S4FS22 E0VHV9 A0A026X2E1 A0A2J7QAL9 A0A2A3EQH2 A0A087U5M2 B4J4Z7 A0A3B0JM64 A0A087ZNU3 A0A3Q0J314 B4HQ06 A0A0J9RCN6 A0A0Q5VN54 B3NRR4 Q7PT15 T1ISC4 A0A1W4VQE7 A0A1B6DJ21 A0A0P5TID5 A0A0P5BB28 A0A0P5AM61 A0A0P5BHA9 B4MPB9 A0A0P5ZNN8 B4LMI5 B4P4U8 B4KSW8 A0A0P5BZN1 A0A1E1X525 A0A0P5ZRJ7 A0A1Q3G110 A0A0P5UF42 A0A0P4VZY0 A0A0P4VUU8 A0A1Q3G109 A0A0N7ZUH2 A0A0P5Y1A6 A0A0P5Z2M3 A0A0P4WEW9 A0A182IUJ2 A0A182QNF6 A0A034V5D9 A0A182KS24 A0A0K8U9Y3 A0A0A1WHQ6 A0A1W4VCC3 A0A0K8UR12 A0A182M8B8 A0A2M4CR01 W5JKS0 A0A2M4CQX7 A0A182RB12
PDB
4HZS
E-value=7.82884e-67,
Score=649
Ontologies
GO
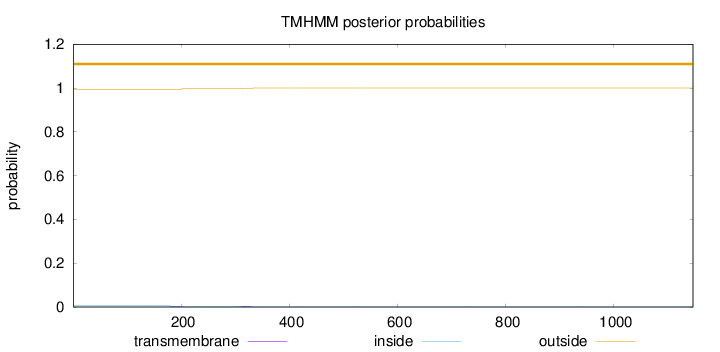
Topology
Length:
1146
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.1241
Exp number, first 60 AAs:
0.00018
Total prob of N-in:
0.00578
outside
1 - 1146
Population Genetic Test Statistics
Pi
18.611621
Theta
17.342894
Tajima's D
-1.329876
CLR
0.024728
CSRT
0.0849957502124894
Interpretation
Uncertain
