Gene
KWMTBOMO00512
Annotation
PREDICTED:_neurocalcin_homolog_[Bombyx_mori]
Full name
Neurocalcin homolog
Location in the cell
Cytoplasmic Reliability : 2.907
Sequence
CDS
ATGGGTAAACAAAATAGTAAACTCAAGCCTGAAGTATTGGAGGATCTGAAACAGAATACGGAGTTTTCAGACGCAGAAATACAAGAATGGTACAAAGGATTCCTGAAGGACTGTCCCAGTGGCCATCTTTCCGTCGAAGAGTTCAAAAAGATATACGGGAATTTTTTCCCTTACGGTGACGCTAGTAAGTTCGCAGAACACGTATTCCGCACATTCGATGCGAACGGCGACGGGACTATAGATTTCAGAGAATTCCTGTGTGCACTTAGTGTGACGTCGCGCGGCAAGCTGGAACAAAAGTTAAAGTGGGCATTCTCCATGTATGACTTAGATGGAAACGGATACATCTCAAGGCAGGAGATGCTGGAAATCGTCACTGCCATTTACAAGATGGTGGGCTCCGTTATGAAGATGCCGGAAGATGAATCTACCCCAGAGAAACGCACCGACAAAATATTCCGTCAGATGGACAGGAACAAGGACGGAAAACTCTCCCTCGAGGAGTTCATAGAAGGAGCCAAGAGCGACCCTTCTATTGTACGGCTGCTCCAATGTGATCCCCAATCTCAGTGA
Protein
MGKQNSKLKPEVLEDLKQNTEFSDAEIQEWYKGFLKDCPSGHLSVEEFKKIYGNFFPYGDASKFAEHVFRTFDANGDGTIDFREFLCALSVTSRGKLEQKLKWAFSMYDLDGNGYISRQEMLEIVTAIYKMVGSVMKMPEDESTPEKRTDKIFRQMDRNKDGKLSLEEFIEGAKSDPSIVRLLQCDPQSQ
Summary
Description
Calcium-binding protein that is essential for promoting night-time sleep and inhibiting nocturnal hyperactivity (PubMed:7989365, PubMed:30865587). During the night-time it promotes sleep by limiting synaptic output from arousal- and wake-promoting neurons within the C01- and A05- domains that include the mushroom body, and the antennal mechanosensory and motor center (PubMed:30865587). Regulated by the circadian clock network and light-sensing circuits that include the CRY photoreceptor; it is likely that both of these internal and external cues are required to define the temporal window (the night-time) during which this protein functions (PubMed:30865587). Inhibits the phosphorylation of rhodopsin in a calcium-dependent manner (PubMed:8626592).
Similarity
Belongs to the recoverin family.
Keywords
Calcium
Complete proteome
Lipoprotein
Metal-binding
Myristate
Reference proteome
Repeat
Feature
chain Neurocalcin homolog
Uniprot
A0A2A4K583
A0A2H1VGM0
A0A212EID6
T1DQE8
A0A1Q3FCV4
U5EWG5
+ More
A0A182YMN6 A0A2M4CSU5 A0A2M3ZBI7 A0A182I2T2 A0A2M4AWJ6 A0A182MSA4 A0A182QW79 A0A182UQ27 A0A182JZ50 B0WLJ1 A0A182WJP8 Q17DD3 A0A182KQR1 A0A182IM38 A0A182FG91 Q7QJ35 A0A182R3H3 A0A182GM84 A0A1J1III6 A0A1L8DM23 A0A1B0DPF3 A0A0K8TP37 A0A1W4W7Y2 A0A1I8M3K6 A0A1A9VEC8 A0A1B0BYY9 B4HBT0 B3M8R6 B4IIU1 B3NE21 B4QRE6 B4IUE1 P42325 A0A2M3ZBN2 D7EI08 A0A1Y1K1J3 A0A0C9RHF7 B4MXN8 A0A0A1WWK3 A0A0K8VXB8 W8AX40 A0A034WTZ2 A0A1B0F9T4 A0A336K594 A0A023EIT5 A0A0M5IYV4 B4J1C5 B4KXP7 K7IP72 A0A0C9RZM1 A0A0T6B8M9 A0A2J7RKX3 A0A2P8YZU5 A0A1I8NM34 A0A151J8X7 F4WKW7 A0A195BJE3 A0A2A3E7E2 V9IMJ2 A0A0M8ZWG3 A0A088AGM2 A0A151K151 E2C1S4 A0A158P3V5 E2AUC2 A0A151WSD4 B4LFN2 A0A1B6EUQ4 A0A1B6H7K7 A0A195CNA8 B7PX38 V5I5H2 A0A1I9WLM4 A0A026VUK0 A0A1B6LTD4 A0A023GI71 A0A224YJ01 A0A131YUR0 L7M547 A0A131XHN9 A0A067RMX2 A0A194QVN3 A0A194PGJ8 A0A1B6D6R2 A0A0F7VK52 A0A1W7RA23 A0A087T645 A0A0P4W9R7 A0A023F5V0 A0A069DQA9 A0A2R5LIB0 A0A2P6KSJ5 T1HKE1 A0A146MHQ1
A0A182YMN6 A0A2M4CSU5 A0A2M3ZBI7 A0A182I2T2 A0A2M4AWJ6 A0A182MSA4 A0A182QW79 A0A182UQ27 A0A182JZ50 B0WLJ1 A0A182WJP8 Q17DD3 A0A182KQR1 A0A182IM38 A0A182FG91 Q7QJ35 A0A182R3H3 A0A182GM84 A0A1J1III6 A0A1L8DM23 A0A1B0DPF3 A0A0K8TP37 A0A1W4W7Y2 A0A1I8M3K6 A0A1A9VEC8 A0A1B0BYY9 B4HBT0 B3M8R6 B4IIU1 B3NE21 B4QRE6 B4IUE1 P42325 A0A2M3ZBN2 D7EI08 A0A1Y1K1J3 A0A0C9RHF7 B4MXN8 A0A0A1WWK3 A0A0K8VXB8 W8AX40 A0A034WTZ2 A0A1B0F9T4 A0A336K594 A0A023EIT5 A0A0M5IYV4 B4J1C5 B4KXP7 K7IP72 A0A0C9RZM1 A0A0T6B8M9 A0A2J7RKX3 A0A2P8YZU5 A0A1I8NM34 A0A151J8X7 F4WKW7 A0A195BJE3 A0A2A3E7E2 V9IMJ2 A0A0M8ZWG3 A0A088AGM2 A0A151K151 E2C1S4 A0A158P3V5 E2AUC2 A0A151WSD4 B4LFN2 A0A1B6EUQ4 A0A1B6H7K7 A0A195CNA8 B7PX38 V5I5H2 A0A1I9WLM4 A0A026VUK0 A0A1B6LTD4 A0A023GI71 A0A224YJ01 A0A131YUR0 L7M547 A0A131XHN9 A0A067RMX2 A0A194QVN3 A0A194PGJ8 A0A1B6D6R2 A0A0F7VK52 A0A1W7RA23 A0A087T645 A0A0P4W9R7 A0A023F5V0 A0A069DQA9 A0A2R5LIB0 A0A2P6KSJ5 T1HKE1 A0A146MHQ1
Pubmed
22118469
25244985
17510324
20966253
12364791
14747013
+ More
17210077 26483478 26369729 25315136 17994087 22936249 17550304 7989365 10731132 12537572 12537569 8626592 30865587 18362917 19820115 28004739 25830018 24495485 25348373 24945155 18057021 20075255 29403074 21719571 20798317 21347285 25765539 27538518 24508170 30249741 28797301 26830274 25576852 28049606 24845553 26354079 25474469 26334808 26823975
17210077 26483478 26369729 25315136 17994087 22936249 17550304 7989365 10731132 12537572 12537569 8626592 30865587 18362917 19820115 28004739 25830018 24495485 25348373 24945155 18057021 20075255 29403074 21719571 20798317 21347285 25765539 27538518 24508170 30249741 28797301 26830274 25576852 28049606 24845553 26354079 25474469 26334808 26823975
EMBL
NWSH01000146
PCG79078.1
ODYU01002454
SOQ39966.1
AGBW02014676
OWR41237.1
+ More
GAMD01002564 JAA99026.1 GFDL01009658 JAV25387.1 GANO01001425 JAB58446.1 GGFL01004219 MBW68397.1 GGFM01005112 MBW25863.1 APCN01000180 GGFK01011846 MBW45167.1 AXCM01006292 AXCN02000543 DS231986 EDS30510.1 CH477297 EAT44357.1 AXCP01006038 AAAB01008807 EAA04175.4 EDO64633.1 EDO64634.1 JXUM01073965 JXUM01073966 KQ562804 KXJ75091.1 CVRI01000054 CRL00045.1 GFDF01006575 JAV07509.1 AJVK01008074 AJVK01008075 GDAI01001692 JAI15911.1 JXJN01022897 CH479267 EDW39524.1 CH902618 EDV41067.2 CH480844 EDW49862.1 CH954178 EDV52445.1 CM000363 CM002912 EDX11159.1 KMZ00671.1 CM000159 CH891888 EDW95243.1 EDX00005.1 U15735 AE014296 AY071612 GGFM01005119 MBW25870.1 DS497669 EFA12929.1 GEZM01095819 GEZM01095818 JAV55324.1 GBYB01012637 JAG82404.1 CH963876 EDW76807.1 GBXI01011060 JAD03232.1 GDHF01024738 GDHF01014793 GDHF01008788 JAI27576.1 JAI37521.1 JAI43526.1 GAMC01013062 GAMC01013061 GAMC01013060 JAB93495.1 GAKP01001293 GAKP01001292 GAKP01001291 JAC57660.1 CCAG010018359 UFQS01000127 UFQT01000127 SSX00146.1 SSX20526.1 GAPW01004732 JAC08866.1 CP012525 ALC44298.1 CH916366 EDV97994.1 CH933809 EDW19754.1 KRG06801.1 GBYB01005054 GBYB01011649 GBYB01014886 GBYB01014895 JAG74821.1 JAG81416.1 JAG84653.1 JAG84662.1 LJIG01009130 KRT83695.1 NEVH01002704 PNF41494.1 PYGN01000263 PSN49758.1 KQ979480 KYN21244.1 GL888206 EGI65209.1 KQ976453 KYM85306.1 KZ288344 PBC27667.1 JR052298 AEY61669.1 KQ435840 KOX71456.1 KQ981292 KYN43126.1 GL451984 EFN78102.1 ADTU01008584 GL442815 EFN62979.1 KQ982783 KYQ50701.1 CH940647 EDW70350.1 KRF84867.1 KRF84868.1 GECZ01028316 JAS41453.1 GECU01037113 GECU01035109 JAS70593.1 JAS72597.1 KQ977513 KYN02198.1 ABJB010718017 DS811420 EEC11160.1 GANP01000360 GEFM01003776 JAB84108.1 JAP72020.1 KU932408 APA34044.1 KK107847 QOIP01000005 EZA47427.1 RLU23136.1 GEBQ01013036 JAT26941.1 GBBM01002690 JAC32728.1 GFPF01006471 MAA17617.1 GEDV01006209 JAP82348.1 GACK01006047 JAA58987.1 GEFH01002866 JAP65715.1 KK852573 KDR21074.1 KQ461108 KPJ09025.1 KQ459605 KPI91834.1 GEDC01031291 GEDC01015924 JAS06007.1 JAS21374.1 LN830691 CFW94200.1 GFAH01000387 JAV48002.1 KK113608 KFM60584.1 GDRN01076412 JAI62897.1 GBBI01002154 JAC16558.1 GBGD01003087 JAC85802.1 GGLE01005103 MBY09229.1 MWRG01005972 PRD29268.1 ACPB03012308 GDHC01013605 GDHC01000353 JAQ05024.1 JAQ18276.1
GAMD01002564 JAA99026.1 GFDL01009658 JAV25387.1 GANO01001425 JAB58446.1 GGFL01004219 MBW68397.1 GGFM01005112 MBW25863.1 APCN01000180 GGFK01011846 MBW45167.1 AXCM01006292 AXCN02000543 DS231986 EDS30510.1 CH477297 EAT44357.1 AXCP01006038 AAAB01008807 EAA04175.4 EDO64633.1 EDO64634.1 JXUM01073965 JXUM01073966 KQ562804 KXJ75091.1 CVRI01000054 CRL00045.1 GFDF01006575 JAV07509.1 AJVK01008074 AJVK01008075 GDAI01001692 JAI15911.1 JXJN01022897 CH479267 EDW39524.1 CH902618 EDV41067.2 CH480844 EDW49862.1 CH954178 EDV52445.1 CM000363 CM002912 EDX11159.1 KMZ00671.1 CM000159 CH891888 EDW95243.1 EDX00005.1 U15735 AE014296 AY071612 GGFM01005119 MBW25870.1 DS497669 EFA12929.1 GEZM01095819 GEZM01095818 JAV55324.1 GBYB01012637 JAG82404.1 CH963876 EDW76807.1 GBXI01011060 JAD03232.1 GDHF01024738 GDHF01014793 GDHF01008788 JAI27576.1 JAI37521.1 JAI43526.1 GAMC01013062 GAMC01013061 GAMC01013060 JAB93495.1 GAKP01001293 GAKP01001292 GAKP01001291 JAC57660.1 CCAG010018359 UFQS01000127 UFQT01000127 SSX00146.1 SSX20526.1 GAPW01004732 JAC08866.1 CP012525 ALC44298.1 CH916366 EDV97994.1 CH933809 EDW19754.1 KRG06801.1 GBYB01005054 GBYB01011649 GBYB01014886 GBYB01014895 JAG74821.1 JAG81416.1 JAG84653.1 JAG84662.1 LJIG01009130 KRT83695.1 NEVH01002704 PNF41494.1 PYGN01000263 PSN49758.1 KQ979480 KYN21244.1 GL888206 EGI65209.1 KQ976453 KYM85306.1 KZ288344 PBC27667.1 JR052298 AEY61669.1 KQ435840 KOX71456.1 KQ981292 KYN43126.1 GL451984 EFN78102.1 ADTU01008584 GL442815 EFN62979.1 KQ982783 KYQ50701.1 CH940647 EDW70350.1 KRF84867.1 KRF84868.1 GECZ01028316 JAS41453.1 GECU01037113 GECU01035109 JAS70593.1 JAS72597.1 KQ977513 KYN02198.1 ABJB010718017 DS811420 EEC11160.1 GANP01000360 GEFM01003776 JAB84108.1 JAP72020.1 KU932408 APA34044.1 KK107847 QOIP01000005 EZA47427.1 RLU23136.1 GEBQ01013036 JAT26941.1 GBBM01002690 JAC32728.1 GFPF01006471 MAA17617.1 GEDV01006209 JAP82348.1 GACK01006047 JAA58987.1 GEFH01002866 JAP65715.1 KK852573 KDR21074.1 KQ461108 KPJ09025.1 KQ459605 KPI91834.1 GEDC01031291 GEDC01015924 JAS06007.1 JAS21374.1 LN830691 CFW94200.1 GFAH01000387 JAV48002.1 KK113608 KFM60584.1 GDRN01076412 JAI62897.1 GBBI01002154 JAC16558.1 GBGD01003087 JAC85802.1 GGLE01005103 MBY09229.1 MWRG01005972 PRD29268.1 ACPB03012308 GDHC01013605 GDHC01000353 JAQ05024.1 JAQ18276.1
Proteomes
UP000218220
UP000007151
UP000076408
UP000075840
UP000075883
UP000075886
+ More
UP000075903 UP000075881 UP000002320 UP000075920 UP000008820 UP000075882 UP000075880 UP000069272 UP000007062 UP000075900 UP000069940 UP000249989 UP000183832 UP000092462 UP000192221 UP000095301 UP000078200 UP000092460 UP000008744 UP000007801 UP000001292 UP000008711 UP000000304 UP000002282 UP000000803 UP000007266 UP000007798 UP000092444 UP000092553 UP000001070 UP000009192 UP000002358 UP000235965 UP000245037 UP000095300 UP000078492 UP000007755 UP000078540 UP000242457 UP000053105 UP000005203 UP000078541 UP000008237 UP000005205 UP000000311 UP000075809 UP000008792 UP000078542 UP000001555 UP000053097 UP000279307 UP000027135 UP000053240 UP000053268 UP000054359 UP000015103
UP000075903 UP000075881 UP000002320 UP000075920 UP000008820 UP000075882 UP000075880 UP000069272 UP000007062 UP000075900 UP000069940 UP000249989 UP000183832 UP000092462 UP000192221 UP000095301 UP000078200 UP000092460 UP000008744 UP000007801 UP000001292 UP000008711 UP000000304 UP000002282 UP000000803 UP000007266 UP000007798 UP000092444 UP000092553 UP000001070 UP000009192 UP000002358 UP000235965 UP000245037 UP000095300 UP000078492 UP000007755 UP000078540 UP000242457 UP000053105 UP000005203 UP000078541 UP000008237 UP000005205 UP000000311 UP000075809 UP000008792 UP000078542 UP000001555 UP000053097 UP000279307 UP000027135 UP000053240 UP000053268 UP000054359 UP000015103
PRIDE
Interpro
SUPFAM
SSF47473
SSF47473
CDD
ProteinModelPortal
A0A2A4K583
A0A2H1VGM0
A0A212EID6
T1DQE8
A0A1Q3FCV4
U5EWG5
+ More
A0A182YMN6 A0A2M4CSU5 A0A2M3ZBI7 A0A182I2T2 A0A2M4AWJ6 A0A182MSA4 A0A182QW79 A0A182UQ27 A0A182JZ50 B0WLJ1 A0A182WJP8 Q17DD3 A0A182KQR1 A0A182IM38 A0A182FG91 Q7QJ35 A0A182R3H3 A0A182GM84 A0A1J1III6 A0A1L8DM23 A0A1B0DPF3 A0A0K8TP37 A0A1W4W7Y2 A0A1I8M3K6 A0A1A9VEC8 A0A1B0BYY9 B4HBT0 B3M8R6 B4IIU1 B3NE21 B4QRE6 B4IUE1 P42325 A0A2M3ZBN2 D7EI08 A0A1Y1K1J3 A0A0C9RHF7 B4MXN8 A0A0A1WWK3 A0A0K8VXB8 W8AX40 A0A034WTZ2 A0A1B0F9T4 A0A336K594 A0A023EIT5 A0A0M5IYV4 B4J1C5 B4KXP7 K7IP72 A0A0C9RZM1 A0A0T6B8M9 A0A2J7RKX3 A0A2P8YZU5 A0A1I8NM34 A0A151J8X7 F4WKW7 A0A195BJE3 A0A2A3E7E2 V9IMJ2 A0A0M8ZWG3 A0A088AGM2 A0A151K151 E2C1S4 A0A158P3V5 E2AUC2 A0A151WSD4 B4LFN2 A0A1B6EUQ4 A0A1B6H7K7 A0A195CNA8 B7PX38 V5I5H2 A0A1I9WLM4 A0A026VUK0 A0A1B6LTD4 A0A023GI71 A0A224YJ01 A0A131YUR0 L7M547 A0A131XHN9 A0A067RMX2 A0A194QVN3 A0A194PGJ8 A0A1B6D6R2 A0A0F7VK52 A0A1W7RA23 A0A087T645 A0A0P4W9R7 A0A023F5V0 A0A069DQA9 A0A2R5LIB0 A0A2P6KSJ5 T1HKE1 A0A146MHQ1
A0A182YMN6 A0A2M4CSU5 A0A2M3ZBI7 A0A182I2T2 A0A2M4AWJ6 A0A182MSA4 A0A182QW79 A0A182UQ27 A0A182JZ50 B0WLJ1 A0A182WJP8 Q17DD3 A0A182KQR1 A0A182IM38 A0A182FG91 Q7QJ35 A0A182R3H3 A0A182GM84 A0A1J1III6 A0A1L8DM23 A0A1B0DPF3 A0A0K8TP37 A0A1W4W7Y2 A0A1I8M3K6 A0A1A9VEC8 A0A1B0BYY9 B4HBT0 B3M8R6 B4IIU1 B3NE21 B4QRE6 B4IUE1 P42325 A0A2M3ZBN2 D7EI08 A0A1Y1K1J3 A0A0C9RHF7 B4MXN8 A0A0A1WWK3 A0A0K8VXB8 W8AX40 A0A034WTZ2 A0A1B0F9T4 A0A336K594 A0A023EIT5 A0A0M5IYV4 B4J1C5 B4KXP7 K7IP72 A0A0C9RZM1 A0A0T6B8M9 A0A2J7RKX3 A0A2P8YZU5 A0A1I8NM34 A0A151J8X7 F4WKW7 A0A195BJE3 A0A2A3E7E2 V9IMJ2 A0A0M8ZWG3 A0A088AGM2 A0A151K151 E2C1S4 A0A158P3V5 E2AUC2 A0A151WSD4 B4LFN2 A0A1B6EUQ4 A0A1B6H7K7 A0A195CNA8 B7PX38 V5I5H2 A0A1I9WLM4 A0A026VUK0 A0A1B6LTD4 A0A023GI71 A0A224YJ01 A0A131YUR0 L7M547 A0A131XHN9 A0A067RMX2 A0A194QVN3 A0A194PGJ8 A0A1B6D6R2 A0A0F7VK52 A0A1W7RA23 A0A087T645 A0A0P4W9R7 A0A023F5V0 A0A069DQA9 A0A2R5LIB0 A0A2P6KSJ5 T1HKE1 A0A146MHQ1
PDB
5T7C
E-value=5.61267e-93,
Score=866
Ontologies
GO
PANTHER
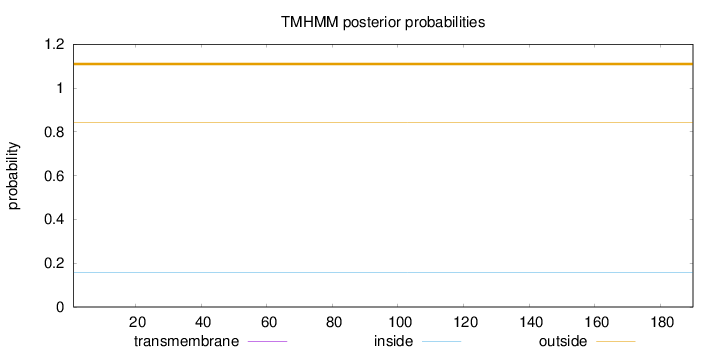
Topology
Length:
190
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00092
Exp number, first 60 AAs:
0
Total prob of N-in:
0.15830
outside
1 - 190
Population Genetic Test Statistics
Pi
17.363152
Theta
18.471115
Tajima's D
-0.551463
CLR
0
CSRT
0.231688415579221
Interpretation
Uncertain
