Gene
KWMTBOMO00508
Pre Gene Modal
BGIBMGA012288
Annotation
PREDICTED:_probable_cationic_amino_acid_transporter_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.969
Sequence
CDS
ATGCAGCGCGTAATGCAGCGTATCCAGCAAGTAGACTGGAAGGCCGTCCGTCGCGACGGTCATAACATCTTCACCAAGCTCATCAGGACGAAGGACCCGTTGGAGGCAGAGGCCAACCAAGAACTGGCTGTGCCGGGGAGACATCGTTTATCGCGTTGTTTAACCACTACGGATCTGACGTCCTTGGGCGTGGGCTCCTGTGTTGGCACCGGGATGTATCTCGTTGCTGGCATGGTGGCCCGCAAGTTCGCAGGCCCTGGCGTTATGATAAGCTTTATCATAGCGGCCATAGCCAGTATCTTTTCGGGTGCATGCTACGCCGAGTTCGGCGTCCGTGTCCCGAATACCACTGGATCGGCTTATACCTACTCATACGTCACCGTTGGAGAGTTCATCGCCTTCATTATTGGTTGGAACATGGTGCTCGAGTACCTGATCGGGACTAGTGCCTGCGCCAGGGCTCTGAGTGCGTGTTTCGACTCTCTCTTCAACGGAGCTATTTCCCACCATCTCGCTCTCAACATCGGAACTATCTTTGGCAAGCCCCCGGACTTCATCGCTTTCGCCATTACTCTTTTGATGATGCTCGTCCTGATCGCTGGCGTGAGGAAATCGCTGATGTTCAACAACATCCTAAACTCCATCAACCTCATAGTTTGGGTGTTCATCATGGGCTCCGGTCTCTTCTACGCCGACTTCAGCAACTGGACTGATCATAAGGGTTTTCTTCCGTACGGATGGAACGGGGTGTTCAGCGGCGCCGCTACTTGCTTCTACGCTTTCATCGGGTTCGATATCATTGCTACCACCGGAGAAGAGGCCAACAACCCTAAGCGTTCCATCCCTCTGGCTATAGTGCTGAGTCTGGGCATCATCCTACTGGCTTATGTTAGCACTTCCATGATGCTGACTCTGATAGTTCCCTACGACCAAATAGACACCGACTCGGCCTTGGTACAAATGTTCGCCCGCGCCGGCGCCCCCATCTGCAAGTACATCGTTGCGATTGGTGCTCTCGCTGGCCTCACCGTGTCCATGTTTGGTTCCATGTTCCCGATGCCAAGGATCATCTATGCAATGGCGCAAGACGGACTAATCTTCAAGTCACTGGCGTCGATTTCGGAGACATCCGGGACTCCAGCCCTCGCTACCATCCTAGGAGGTTTCGCTGCCGCAATGGTAGCCCTGACTATCCAGCTGGAGGTGCTCGTTGAAATGATGAGCATCGTTTCTGCAGCTATGTCAAACTCAAAAACAACTCAAACAACAGCAGTCGAGATGCTCTGGAACGAAACATCCAGAACAGACGCTAGTTAG
Protein
MQRVMQRIQQVDWKAVRRDGHNIFTKLIRTKDPLEAEANQELAVPGRHRLSRCLTTTDLTSLGVGSCVGTGMYLVAGMVARKFAGPGVMISFIIAAIASIFSGACYAEFGVRVPNTTGSAYTYSYVTVGEFIAFIIGWNMVLEYLIGTSACARALSACFDSLFNGAISHHLALNIGTIFGKPPDFIAFAITLLMMLVLIAGVRKSLMFNNILNSINLIVWVFIMGSGLFYADFSNWTDHKGFLPYGWNGVFSGAATCFYAFIGFDIIATTGEEANNPKRSIPLAIVLSLGIILLAYVSTSMMLTLIVPYDQIDTDSALVQMFARAGAPICKYIVAIGALAGLTVSMFGSMFPMPRIIYAMAQDGLIFKSLASISETSGTPALATILGGFAAAMVALTIQLEVLVEMMSIVSAAMSNSKTTQTTAVEMLWNETSRTDAS
Summary
Uniprot
A0A212EID0
A0A194QZJ9
A0A194PGK3
A0A195FVI2
A0A195DCR8
A0A232EHE3
+ More
A0A151X4L8 K7IMD9 A0A195BCE8 A0A158NS32 A0A087ZQV8 E2AYK4 A0A2J7RR55 D6WJP6 A0A151IL35 A0A026W979 A0A0C9QGS6 A0A067RNH5 A0A154PAI5 T1IVD4 A0A2A3E3S9 A0A1W4WGI8 N6TKK8 A0A0R3NZF1 A0A084W0L1 A0A182NQN5 Q16PF2 A0A182HKF4 A0A182J0S5 A0A182QIV3 A0A0A1WNX4 A0A182L8N8 A0A182X9H0 Q7PUC7 B4H4Q1 A0A182VND0 A0A226ELU3 A0A182VVX1 G7H832 A0A182PRR9 A0A1J1J420 A0A1I8M5B6 A0A0Q9XG06 A0A1I8P3R8 B3MQM8 Q29HX8 B4I7B1 A0A1B0CEC9 A0A1W4VHJ8 Q9VWD3 A0A3B0KJP4 B3NVT3 A0A0Q5T6Y3 B4PZ08 M9PHM0 B0W2E1 A0A0R1EB45 W8BK65 B4NQ70 B4MAR7 B4L804 A0A0M5J3B6 A0A165RBA4 A0A2H8TZS3 B4JJI0 A0A2R5L4A6 J9KAK6 A0A2S2PER9 A0A1Y1KQG9 A0A182RA48 A0A182FSA7 E0VXE5 A0A1A9ZQI7 A0A1B0FI77 L7M0D3 A0A1A9USW9 A0A1B0C7E0 T1I6V8 A0A0L7R0X5 T1KX13 A0A1A9XCV1 A0A1S3IBV3 A0A3P8S2D7 A0A3B5APM9 A0A1Y1KVD8 W5KFW3 A0A2D0PW17 A0A310TMS3 H3CMF8 A0A3B4CU92 A0A1L8GAM3 A0A3B1JR23 Q4STY8 A0A3L8DB57 A0A3N0YEI9 W5MKB5 A0A2K5DW10 A0A0S7HYA8
A0A151X4L8 K7IMD9 A0A195BCE8 A0A158NS32 A0A087ZQV8 E2AYK4 A0A2J7RR55 D6WJP6 A0A151IL35 A0A026W979 A0A0C9QGS6 A0A067RNH5 A0A154PAI5 T1IVD4 A0A2A3E3S9 A0A1W4WGI8 N6TKK8 A0A0R3NZF1 A0A084W0L1 A0A182NQN5 Q16PF2 A0A182HKF4 A0A182J0S5 A0A182QIV3 A0A0A1WNX4 A0A182L8N8 A0A182X9H0 Q7PUC7 B4H4Q1 A0A182VND0 A0A226ELU3 A0A182VVX1 G7H832 A0A182PRR9 A0A1J1J420 A0A1I8M5B6 A0A0Q9XG06 A0A1I8P3R8 B3MQM8 Q29HX8 B4I7B1 A0A1B0CEC9 A0A1W4VHJ8 Q9VWD3 A0A3B0KJP4 B3NVT3 A0A0Q5T6Y3 B4PZ08 M9PHM0 B0W2E1 A0A0R1EB45 W8BK65 B4NQ70 B4MAR7 B4L804 A0A0M5J3B6 A0A165RBA4 A0A2H8TZS3 B4JJI0 A0A2R5L4A6 J9KAK6 A0A2S2PER9 A0A1Y1KQG9 A0A182RA48 A0A182FSA7 E0VXE5 A0A1A9ZQI7 A0A1B0FI77 L7M0D3 A0A1A9USW9 A0A1B0C7E0 T1I6V8 A0A0L7R0X5 T1KX13 A0A1A9XCV1 A0A1S3IBV3 A0A3P8S2D7 A0A3B5APM9 A0A1Y1KVD8 W5KFW3 A0A2D0PW17 A0A310TMS3 H3CMF8 A0A3B4CU92 A0A1L8GAM3 A0A3B1JR23 Q4STY8 A0A3L8DB57 A0A3N0YEI9 W5MKB5 A0A2K5DW10 A0A0S7HYA8
Pubmed
22118469
26354079
28648823
20075255
21347285
20798317
+ More
18362917 19820115 24508170 24845553 23537049 15632085 17994087 24438588 17510324 25830018 20966253 12364791 14747013 17210077 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 24495485 18057021 28004739 20566863 25576852 25329095 15496914 27762356 30249741
18362917 19820115 24508170 24845553 23537049 15632085 17994087 24438588 17510324 25830018 20966253 12364791 14747013 17210077 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17550304 24495485 18057021 28004739 20566863 25576852 25329095 15496914 27762356 30249741
EMBL
AGBW02014676
OWR41234.1
KQ461108
KPJ09021.1
KQ459605
KPI91839.1
+ More
KQ981208 KYN44650.1 KQ980989 KYN10705.1 NNAY01004518 OXU17780.1 KQ982544 KYQ55352.1 KQ976528 KYM81870.1 ADTU01024436 ADTU01024437 ADTU01024438 ADTU01024439 GL443918 EFN61492.1 NEVH01000611 PNF43297.1 KQ971343 EFA04475.1 KQ977142 KYN05322.1 KK107374 EZA51589.1 GBYB01013773 GBYB01013774 JAG83540.1 JAG83541.1 KK852521 KDR22125.1 KQ434857 KZC08862.1 JH431581 KZ288439 PBC25741.1 APGK01034130 APGK01034131 KB740904 KB632006 ENN78458.1 ERL87969.1 CH379064 KRT06408.1 ATLV01019122 KE525262 KFB43755.1 CH477784 EAT36248.1 APCN01000826 AXCN02001509 GBXI01014164 GBXI01005267 JAD00128.1 JAD09025.1 AAAB01008987 EAA01745.5 CH479209 EDW32663.1 LNIX01000003 OXA58117.1 BT132755 AET07638.1 CVRI01000070 CRL07152.1 CH933814 KRG07456.1 CH902621 EDV44654.1 EAL31629.2 CH480823 EDW56209.1 AJWK01008736 AE014298 AAF49012.2 OUUW01000011 SPP86709.1 CH954180 EDV46748.1 KQS30104.1 CM000162 EDX02086.1 AGB95568.1 DS231826 EDS28687.1 KRK06557.1 GAMC01007438 JAB99117.1 CH964291 EDW86295.1 CH940655 EDW66326.2 EDW05579.1 KRG07454.1 KRG07455.1 KRG07457.1 CP012528 ALC49561.1 KR028430 AML23856.1 GFXV01007962 MBW19767.1 CH916370 EDV99732.1 GGLE01000202 MBY04328.1 ABLF02029856 ABLF02029858 ABLF02029859 ABLF02029860 GGMR01015330 MBY27949.1 GEZM01076756 GEZM01076753 JAV63669.1 DS235830 EEB18051.1 CCAG010004711 GACK01008466 JAA56568.1 JXJN01028274 JXJN01028275 ACPB03000878 KQ414670 KOC64446.1 CAEY01000675 GEZM01076751 JAV63675.1 KV467268 OCT56514.1 CM004474 OCT80776.1 CAAE01014066 CAF95894.1 QOIP01000010 RLU17730.1 RJVU01044706 ROL44665.1 AHAT01015260 AHAT01015261 GBYX01435555 JAO45780.1
KQ981208 KYN44650.1 KQ980989 KYN10705.1 NNAY01004518 OXU17780.1 KQ982544 KYQ55352.1 KQ976528 KYM81870.1 ADTU01024436 ADTU01024437 ADTU01024438 ADTU01024439 GL443918 EFN61492.1 NEVH01000611 PNF43297.1 KQ971343 EFA04475.1 KQ977142 KYN05322.1 KK107374 EZA51589.1 GBYB01013773 GBYB01013774 JAG83540.1 JAG83541.1 KK852521 KDR22125.1 KQ434857 KZC08862.1 JH431581 KZ288439 PBC25741.1 APGK01034130 APGK01034131 KB740904 KB632006 ENN78458.1 ERL87969.1 CH379064 KRT06408.1 ATLV01019122 KE525262 KFB43755.1 CH477784 EAT36248.1 APCN01000826 AXCN02001509 GBXI01014164 GBXI01005267 JAD00128.1 JAD09025.1 AAAB01008987 EAA01745.5 CH479209 EDW32663.1 LNIX01000003 OXA58117.1 BT132755 AET07638.1 CVRI01000070 CRL07152.1 CH933814 KRG07456.1 CH902621 EDV44654.1 EAL31629.2 CH480823 EDW56209.1 AJWK01008736 AE014298 AAF49012.2 OUUW01000011 SPP86709.1 CH954180 EDV46748.1 KQS30104.1 CM000162 EDX02086.1 AGB95568.1 DS231826 EDS28687.1 KRK06557.1 GAMC01007438 JAB99117.1 CH964291 EDW86295.1 CH940655 EDW66326.2 EDW05579.1 KRG07454.1 KRG07455.1 KRG07457.1 CP012528 ALC49561.1 KR028430 AML23856.1 GFXV01007962 MBW19767.1 CH916370 EDV99732.1 GGLE01000202 MBY04328.1 ABLF02029856 ABLF02029858 ABLF02029859 ABLF02029860 GGMR01015330 MBY27949.1 GEZM01076756 GEZM01076753 JAV63669.1 DS235830 EEB18051.1 CCAG010004711 GACK01008466 JAA56568.1 JXJN01028274 JXJN01028275 ACPB03000878 KQ414670 KOC64446.1 CAEY01000675 GEZM01076751 JAV63675.1 KV467268 OCT56514.1 CM004474 OCT80776.1 CAAE01014066 CAF95894.1 QOIP01000010 RLU17730.1 RJVU01044706 ROL44665.1 AHAT01015260 AHAT01015261 GBYX01435555 JAO45780.1
Proteomes
UP000007151
UP000053240
UP000053268
UP000078541
UP000078492
UP000215335
+ More
UP000075809 UP000002358 UP000078540 UP000005205 UP000005203 UP000000311 UP000235965 UP000007266 UP000078542 UP000053097 UP000027135 UP000076502 UP000242457 UP000192223 UP000019118 UP000030742 UP000001819 UP000030765 UP000075884 UP000008820 UP000075840 UP000075880 UP000075886 UP000075882 UP000076407 UP000007062 UP000008744 UP000075903 UP000198287 UP000075920 UP000075885 UP000183832 UP000095301 UP000009192 UP000095300 UP000007801 UP000001292 UP000092461 UP000192221 UP000000803 UP000268350 UP000008711 UP000002282 UP000002320 UP000007798 UP000008792 UP000092553 UP000001070 UP000007819 UP000075900 UP000069272 UP000009046 UP000092445 UP000092444 UP000078200 UP000092460 UP000015103 UP000053825 UP000015104 UP000092443 UP000085678 UP000265080 UP000261400 UP000018467 UP000221080 UP000007303 UP000261440 UP000186698 UP000279307 UP000018468 UP000233020
UP000075809 UP000002358 UP000078540 UP000005205 UP000005203 UP000000311 UP000235965 UP000007266 UP000078542 UP000053097 UP000027135 UP000076502 UP000242457 UP000192223 UP000019118 UP000030742 UP000001819 UP000030765 UP000075884 UP000008820 UP000075840 UP000075880 UP000075886 UP000075882 UP000076407 UP000007062 UP000008744 UP000075903 UP000198287 UP000075920 UP000075885 UP000183832 UP000095301 UP000009192 UP000095300 UP000007801 UP000001292 UP000092461 UP000192221 UP000000803 UP000268350 UP000008711 UP000002282 UP000002320 UP000007798 UP000008792 UP000092553 UP000001070 UP000007819 UP000075900 UP000069272 UP000009046 UP000092445 UP000092444 UP000078200 UP000092460 UP000015103 UP000053825 UP000015104 UP000092443 UP000085678 UP000265080 UP000261400 UP000018467 UP000221080 UP000007303 UP000261440 UP000186698 UP000279307 UP000018468 UP000233020
ProteinModelPortal
A0A212EID0
A0A194QZJ9
A0A194PGK3
A0A195FVI2
A0A195DCR8
A0A232EHE3
+ More
A0A151X4L8 K7IMD9 A0A195BCE8 A0A158NS32 A0A087ZQV8 E2AYK4 A0A2J7RR55 D6WJP6 A0A151IL35 A0A026W979 A0A0C9QGS6 A0A067RNH5 A0A154PAI5 T1IVD4 A0A2A3E3S9 A0A1W4WGI8 N6TKK8 A0A0R3NZF1 A0A084W0L1 A0A182NQN5 Q16PF2 A0A182HKF4 A0A182J0S5 A0A182QIV3 A0A0A1WNX4 A0A182L8N8 A0A182X9H0 Q7PUC7 B4H4Q1 A0A182VND0 A0A226ELU3 A0A182VVX1 G7H832 A0A182PRR9 A0A1J1J420 A0A1I8M5B6 A0A0Q9XG06 A0A1I8P3R8 B3MQM8 Q29HX8 B4I7B1 A0A1B0CEC9 A0A1W4VHJ8 Q9VWD3 A0A3B0KJP4 B3NVT3 A0A0Q5T6Y3 B4PZ08 M9PHM0 B0W2E1 A0A0R1EB45 W8BK65 B4NQ70 B4MAR7 B4L804 A0A0M5J3B6 A0A165RBA4 A0A2H8TZS3 B4JJI0 A0A2R5L4A6 J9KAK6 A0A2S2PER9 A0A1Y1KQG9 A0A182RA48 A0A182FSA7 E0VXE5 A0A1A9ZQI7 A0A1B0FI77 L7M0D3 A0A1A9USW9 A0A1B0C7E0 T1I6V8 A0A0L7R0X5 T1KX13 A0A1A9XCV1 A0A1S3IBV3 A0A3P8S2D7 A0A3B5APM9 A0A1Y1KVD8 W5KFW3 A0A2D0PW17 A0A310TMS3 H3CMF8 A0A3B4CU92 A0A1L8GAM3 A0A3B1JR23 Q4STY8 A0A3L8DB57 A0A3N0YEI9 W5MKB5 A0A2K5DW10 A0A0S7HYA8
A0A151X4L8 K7IMD9 A0A195BCE8 A0A158NS32 A0A087ZQV8 E2AYK4 A0A2J7RR55 D6WJP6 A0A151IL35 A0A026W979 A0A0C9QGS6 A0A067RNH5 A0A154PAI5 T1IVD4 A0A2A3E3S9 A0A1W4WGI8 N6TKK8 A0A0R3NZF1 A0A084W0L1 A0A182NQN5 Q16PF2 A0A182HKF4 A0A182J0S5 A0A182QIV3 A0A0A1WNX4 A0A182L8N8 A0A182X9H0 Q7PUC7 B4H4Q1 A0A182VND0 A0A226ELU3 A0A182VVX1 G7H832 A0A182PRR9 A0A1J1J420 A0A1I8M5B6 A0A0Q9XG06 A0A1I8P3R8 B3MQM8 Q29HX8 B4I7B1 A0A1B0CEC9 A0A1W4VHJ8 Q9VWD3 A0A3B0KJP4 B3NVT3 A0A0Q5T6Y3 B4PZ08 M9PHM0 B0W2E1 A0A0R1EB45 W8BK65 B4NQ70 B4MAR7 B4L804 A0A0M5J3B6 A0A165RBA4 A0A2H8TZS3 B4JJI0 A0A2R5L4A6 J9KAK6 A0A2S2PER9 A0A1Y1KQG9 A0A182RA48 A0A182FSA7 E0VXE5 A0A1A9ZQI7 A0A1B0FI77 L7M0D3 A0A1A9USW9 A0A1B0C7E0 T1I6V8 A0A0L7R0X5 T1KX13 A0A1A9XCV1 A0A1S3IBV3 A0A3P8S2D7 A0A3B5APM9 A0A1Y1KVD8 W5KFW3 A0A2D0PW17 A0A310TMS3 H3CMF8 A0A3B4CU92 A0A1L8GAM3 A0A3B1JR23 Q4STY8 A0A3L8DB57 A0A3N0YEI9 W5MKB5 A0A2K5DW10 A0A0S7HYA8
PDB
6F34
E-value=1.23262e-51,
Score=514
Ontologies
GO
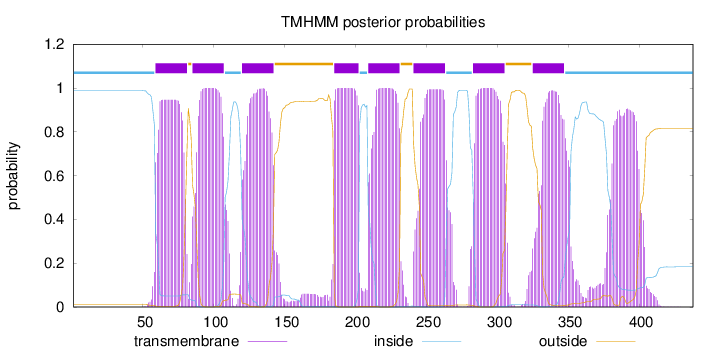
Topology
Length:
438
Number of predicted TMHs:
8
Exp number of AAs in TMHs:
195.87955
Exp number, first 60 AAs:
1.70951
Total prob of N-in:
0.98882
inside
1 - 58
TMhelix
59 - 81
outside
82 - 84
TMhelix
85 - 107
inside
108 - 119
TMhelix
120 - 142
outside
143 - 184
TMhelix
185 - 202
inside
203 - 208
TMhelix
209 - 231
outside
232 - 240
TMhelix
241 - 263
inside
264 - 282
TMhelix
283 - 305
outside
306 - 324
TMhelix
325 - 347
inside
348 - 438
Population Genetic Test Statistics
Pi
19.324333
Theta
20.428148
Tajima's D
-1.369282
CLR
0.033222
CSRT
0.0735463226838658
Interpretation
Uncertain
