Gene
KWMTBOMO00505
Pre Gene Modal
BGIBMGA012289
Annotation
PREDICTED:_uncharacterized_protein_LOC107172728_[Diuraphis_noxia]
Location in the cell
Nuclear Reliability : 2.484
Sequence
CDS
ATGCGTAAAATACTCATAGAACTTGGGTACAAGTACATACAAGCTTCATATGGTCGTAAACAATTGATGGAAAGGACTGACATCGTTTGTGCGCGGCTACAATTCCTGAGAAAACTCAAGACCCTGAGAGATAGTGAAGACACTCGACCAAGAATTTATTTAGATGAGACTTGGGTGAACCAAAATCACAGCCGCAAACGAATTTGGCTAGACAAGGATGGACAAGGTGGCTTAAATACAAAAACGGAAAAGAGGGGCAAGCTAATTGTTCGTCATGCAGGCTGCGCAAAGTATGGTTTTATACCCGAAGCCAGATGGGTCTTCCGCGCTGTGAAGTCTCCGAACGAGGGCTACCATTCAGAAATGAATGCCAAAGGCTTCACACACTGGTTTACAAAACTGCTGCAGAGTCTTGAAGAACTATCAGTCATCATAATGGACAACGCACCCTACCACTCCCAGCAAATAAATCAGGCACCAACATCAGCTAACAAAAAGAGTGAAATTCAGGAGTGGCTCCGCGAACACGGGATTGAGTTTAGTGAGAGTGAGCTCAAGCCGGAACTATTACAAAAGGTGCACCGGAACCGACCAGAAAAAATATACGAATTGTATCAGTTAGCTTTGGAGTCAGGACACGAAGTGATTCGCTTACCACCTTACCATTGCCAATATAACCCAATTGAGTTGGTATGGGCTCAAGCAAAAAATGAAGTTGCCAGCAAAAATAGAACTTTTAAAATTGCTGACGTCGAAGTGTTAGTGCATAAGGCAATTTCGAACGTTTCGCAAAAGAACTGGGAAGATTGTGTGAAGCATGCAGAAAAACTCCAAGAGGTAGACTTGAAGAAATCTCATCTGGCAGAAGAATTCGTAATTCATGTGGATGACGATGACTCTGATGATCCTGATTTATTAGTATACTAA
Protein
MRKILIELGYKYIQASYGRKQLMERTDIVCARLQFLRKLKTLRDSEDTRPRIYLDETWVNQNHSRKRIWLDKDGQGGLNTKTEKRGKLIVRHAGCAKYGFIPEARWVFRAVKSPNEGYHSEMNAKGFTHWFTKLLQSLEELSVIIMDNAPYHSQQINQAPTSANKKSEIQEWLREHGIEFSESELKPELLQKVHRNRPEKIYELYQLALESGHEVIRLPPYHCQYNPIELVWAQAKNEVASKNRTFKIADVEVLVHKAISNVSQKNWEDCVKHAEKLQEVDLKKSHLAEEFVIHVDDDDSDDPDLLVY
Summary
Uniprot
H9JRX9
A0A2S2NY59
J9M601
A0A2S2QN01
A0A2S2PH93
A0A2P8XEU9
+ More
X1XM98 A0A2S2PM81 A0A2S2NB48 A0A2S2P4Q5 J9L3P3 A0A2S2P977 A0A2S2NGI9 A0A2S2PXB9 A0A1E1XJ56 A0A1S3DLW8 J9KMB7 A0A0P4W325 A0A2H1VS09 A0A2A4K5N3 A0A1E1XIJ4 A0A1Y1JVR5 A0A1Y1K3F5 A0A1E1XNA2 A0A1Y1K136 A0A1Y1JYU9 V5HNM0 A0A0B7BSV4 A0A131YB14 A0A0B7BSL6 A0A087TFM8 A0A0B7BUC3 J9LFZ2 H9JUW8 X1XDY6 A0A2P8YMB9 A0A226F1W3 J9KRA4 J9LKT7 A0A2S2R4W8 J9LRK7 X1XNU8 A0A2P8XDU4 A0A1Y1MEN8 X1X3Z5 X1XLE5 V5I8I1 A0A026W2S1 X1WST5 A0A2J7Q5G4 A0A2R7WV07 X1XN91 A0A067QLI2 A0A2P8YXG2 J9LJT6 J9LJ01 A0A2S2R642 A0A2S2QX80 A0A2S2PKU7 A0A0C9QBV3 J9K7A5 A0A2S2PVM9 J9M563 A0A1B6M3D7 J9JPS5 A0A2S2NBG4 J9JSC0 A0A2P8YFT4 J9JW56 A0A2W1B072 A0A1H6FE09 A0A2S2N8L4 A0A226CY92 A0A2S2PF27 A0A3C1TPI2 D2A336 J9K8H7 J9M5W4 A0A2S2QDI3 J9L8L7 X1XDK5 A0A2H1X3K3 D7EKQ4 A0A226D544 A0A087T1E7 J9M9H5 J9M126 Q49HI7
X1XM98 A0A2S2PM81 A0A2S2NB48 A0A2S2P4Q5 J9L3P3 A0A2S2P977 A0A2S2NGI9 A0A2S2PXB9 A0A1E1XJ56 A0A1S3DLW8 J9KMB7 A0A0P4W325 A0A2H1VS09 A0A2A4K5N3 A0A1E1XIJ4 A0A1Y1JVR5 A0A1Y1K3F5 A0A1E1XNA2 A0A1Y1K136 A0A1Y1JYU9 V5HNM0 A0A0B7BSV4 A0A131YB14 A0A0B7BSL6 A0A087TFM8 A0A0B7BUC3 J9LFZ2 H9JUW8 X1XDY6 A0A2P8YMB9 A0A226F1W3 J9KRA4 J9LKT7 A0A2S2R4W8 J9LRK7 X1XNU8 A0A2P8XDU4 A0A1Y1MEN8 X1X3Z5 X1XLE5 V5I8I1 A0A026W2S1 X1WST5 A0A2J7Q5G4 A0A2R7WV07 X1XN91 A0A067QLI2 A0A2P8YXG2 J9LJT6 J9LJ01 A0A2S2R642 A0A2S2QX80 A0A2S2PKU7 A0A0C9QBV3 J9K7A5 A0A2S2PVM9 J9M563 A0A1B6M3D7 J9JPS5 A0A2S2NBG4 J9JSC0 A0A2P8YFT4 J9JW56 A0A2W1B072 A0A1H6FE09 A0A2S2N8L4 A0A226CY92 A0A2S2PF27 A0A3C1TPI2 D2A336 J9K8H7 J9M5W4 A0A2S2QDI3 J9L8L7 X1XDK5 A0A2H1X3K3 D7EKQ4 A0A226D544 A0A087T1E7 J9M9H5 J9M126 Q49HI7
Pubmed
EMBL
BABH01016974
GGMR01009279
MBY21898.1
ABLF02012070
GGMS01009946
MBY79149.1
+ More
GGMR01016203 MBY28822.1 PYGN01002415 PSN30531.1 ABLF02016308 GGMR01017868 MBY30487.1 GGMR01001387 MBY14006.1 GGMR01011267 MBY23886.1 ABLF02031390 GGMR01013129 MBY25748.1 GGMR01003609 MBY16228.1 GGMS01000992 MBY70195.1 GFAC01000052 JAT99136.1 ABLF02005303 GDRN01091729 GDRN01091726 JAI60230.1 ODYU01004103 SOQ43625.1 NWSH01000101 PCG79555.1 GFAC01000066 JAT99122.1 GEZM01099244 JAV53412.1 GEZM01099242 GEZM01099238 JAV53417.1 GFAA01002676 JAU00759.1 GEZM01099240 JAV53415.1 GEZM01099245 GEZM01099237 JAV53411.1 GANP01007266 JAB77202.1 HACG01049198 HACG01049200 CEK96063.1 CEK96065.1 GEFM01000520 JAP75276.1 HACG01049078 CEK95943.1 KK114998 KFM63917.1 HACG01049081 CEK95946.1 ABLF02039436 ABLF02039447 ABLF02066873 BABH01039803 ABLF02064933 PYGN01000495 PSN45395.1 LNIX01000001 OXA63458.1 ABLF02010686 ABLF02032398 GGMS01015884 MBY85087.1 ABLF02006655 ABLF02009920 PYGN01002624 PSN30176.1 GEZM01034973 GEZM01034971 GEZM01034970 GEZM01034965 GEZM01034964 GEZM01034962 GEZM01034955 GEZM01034953 JAV83478.1 ABLF02042318 ABLF02066501 ABLF02037317 GALX01004645 JAB63821.1 KK107522 EZA49329.1 ABLF02011197 ABLF02011198 ABLF02011199 ABLF02042210 NEVH01017562 PNF23820.1 KK855644 PTY23394.1 ABLF02009895 KK853287 KDR08934.1 PYGN01000302 PSN48936.1 ABLF02066315 ABLF02023303 GGMS01016324 MBY85527.1 GGMS01013166 MBY82369.1 GGMR01017451 MBY30070.1 GBYB01011953 JAG81720.1 ABLF02038234 GGMR01020297 MBY32916.1 ABLF02035127 GEBQ01009535 JAT30442.1 ABLF02037452 GGMR01001497 MBY14116.1 ABLF02031602 ABLF02031606 PYGN01000629 PSN43112.1 ABLF02007992 KZ150527 PZC70602.1 FMSV02000534 SEH07569.1 GGMR01000898 MBY13517.1 LNIX01000051 OXA37923.1 GGMR01015375 MBY27994.1 DMSM01000084 HAO52838.1 KQ971339 EFA02787.1 ABLF02025022 ABLF02003450 ABLF02005306 GGMS01006586 MBY75789.1 ABLF02039881 ABLF02018828 ABLF02043063 ODYU01013164 SOQ59857.1 DS497765 EFA11630.1 LNIX01000032 OXA40685.1 KK112951 KFM58936.1 ABLF02006579 ABLF02013842 ABLF02013846 AY907538 AAY00050.1
GGMR01016203 MBY28822.1 PYGN01002415 PSN30531.1 ABLF02016308 GGMR01017868 MBY30487.1 GGMR01001387 MBY14006.1 GGMR01011267 MBY23886.1 ABLF02031390 GGMR01013129 MBY25748.1 GGMR01003609 MBY16228.1 GGMS01000992 MBY70195.1 GFAC01000052 JAT99136.1 ABLF02005303 GDRN01091729 GDRN01091726 JAI60230.1 ODYU01004103 SOQ43625.1 NWSH01000101 PCG79555.1 GFAC01000066 JAT99122.1 GEZM01099244 JAV53412.1 GEZM01099242 GEZM01099238 JAV53417.1 GFAA01002676 JAU00759.1 GEZM01099240 JAV53415.1 GEZM01099245 GEZM01099237 JAV53411.1 GANP01007266 JAB77202.1 HACG01049198 HACG01049200 CEK96063.1 CEK96065.1 GEFM01000520 JAP75276.1 HACG01049078 CEK95943.1 KK114998 KFM63917.1 HACG01049081 CEK95946.1 ABLF02039436 ABLF02039447 ABLF02066873 BABH01039803 ABLF02064933 PYGN01000495 PSN45395.1 LNIX01000001 OXA63458.1 ABLF02010686 ABLF02032398 GGMS01015884 MBY85087.1 ABLF02006655 ABLF02009920 PYGN01002624 PSN30176.1 GEZM01034973 GEZM01034971 GEZM01034970 GEZM01034965 GEZM01034964 GEZM01034962 GEZM01034955 GEZM01034953 JAV83478.1 ABLF02042318 ABLF02066501 ABLF02037317 GALX01004645 JAB63821.1 KK107522 EZA49329.1 ABLF02011197 ABLF02011198 ABLF02011199 ABLF02042210 NEVH01017562 PNF23820.1 KK855644 PTY23394.1 ABLF02009895 KK853287 KDR08934.1 PYGN01000302 PSN48936.1 ABLF02066315 ABLF02023303 GGMS01016324 MBY85527.1 GGMS01013166 MBY82369.1 GGMR01017451 MBY30070.1 GBYB01011953 JAG81720.1 ABLF02038234 GGMR01020297 MBY32916.1 ABLF02035127 GEBQ01009535 JAT30442.1 ABLF02037452 GGMR01001497 MBY14116.1 ABLF02031602 ABLF02031606 PYGN01000629 PSN43112.1 ABLF02007992 KZ150527 PZC70602.1 FMSV02000534 SEH07569.1 GGMR01000898 MBY13517.1 LNIX01000051 OXA37923.1 GGMR01015375 MBY27994.1 DMSM01000084 HAO52838.1 KQ971339 EFA02787.1 ABLF02025022 ABLF02003450 ABLF02005306 GGMS01006586 MBY75789.1 ABLF02039881 ABLF02018828 ABLF02043063 ODYU01013164 SOQ59857.1 DS497765 EFA11630.1 LNIX01000032 OXA40685.1 KK112951 KFM58936.1 ABLF02006579 ABLF02013842 ABLF02013846 AY907538 AAY00050.1
Proteomes
Interpro
SUPFAM
SSF46689
SSF46689
ProteinModelPortal
H9JRX9
A0A2S2NY59
J9M601
A0A2S2QN01
A0A2S2PH93
A0A2P8XEU9
+ More
X1XM98 A0A2S2PM81 A0A2S2NB48 A0A2S2P4Q5 J9L3P3 A0A2S2P977 A0A2S2NGI9 A0A2S2PXB9 A0A1E1XJ56 A0A1S3DLW8 J9KMB7 A0A0P4W325 A0A2H1VS09 A0A2A4K5N3 A0A1E1XIJ4 A0A1Y1JVR5 A0A1Y1K3F5 A0A1E1XNA2 A0A1Y1K136 A0A1Y1JYU9 V5HNM0 A0A0B7BSV4 A0A131YB14 A0A0B7BSL6 A0A087TFM8 A0A0B7BUC3 J9LFZ2 H9JUW8 X1XDY6 A0A2P8YMB9 A0A226F1W3 J9KRA4 J9LKT7 A0A2S2R4W8 J9LRK7 X1XNU8 A0A2P8XDU4 A0A1Y1MEN8 X1X3Z5 X1XLE5 V5I8I1 A0A026W2S1 X1WST5 A0A2J7Q5G4 A0A2R7WV07 X1XN91 A0A067QLI2 A0A2P8YXG2 J9LJT6 J9LJ01 A0A2S2R642 A0A2S2QX80 A0A2S2PKU7 A0A0C9QBV3 J9K7A5 A0A2S2PVM9 J9M563 A0A1B6M3D7 J9JPS5 A0A2S2NBG4 J9JSC0 A0A2P8YFT4 J9JW56 A0A2W1B072 A0A1H6FE09 A0A2S2N8L4 A0A226CY92 A0A2S2PF27 A0A3C1TPI2 D2A336 J9K8H7 J9M5W4 A0A2S2QDI3 J9L8L7 X1XDK5 A0A2H1X3K3 D7EKQ4 A0A226D544 A0A087T1E7 J9M9H5 J9M126 Q49HI7
X1XM98 A0A2S2PM81 A0A2S2NB48 A0A2S2P4Q5 J9L3P3 A0A2S2P977 A0A2S2NGI9 A0A2S2PXB9 A0A1E1XJ56 A0A1S3DLW8 J9KMB7 A0A0P4W325 A0A2H1VS09 A0A2A4K5N3 A0A1E1XIJ4 A0A1Y1JVR5 A0A1Y1K3F5 A0A1E1XNA2 A0A1Y1K136 A0A1Y1JYU9 V5HNM0 A0A0B7BSV4 A0A131YB14 A0A0B7BSL6 A0A087TFM8 A0A0B7BUC3 J9LFZ2 H9JUW8 X1XDY6 A0A2P8YMB9 A0A226F1W3 J9KRA4 J9LKT7 A0A2S2R4W8 J9LRK7 X1XNU8 A0A2P8XDU4 A0A1Y1MEN8 X1X3Z5 X1XLE5 V5I8I1 A0A026W2S1 X1WST5 A0A2J7Q5G4 A0A2R7WV07 X1XN91 A0A067QLI2 A0A2P8YXG2 J9LJT6 J9LJ01 A0A2S2R642 A0A2S2QX80 A0A2S2PKU7 A0A0C9QBV3 J9K7A5 A0A2S2PVM9 J9M563 A0A1B6M3D7 J9JPS5 A0A2S2NBG4 J9JSC0 A0A2P8YFT4 J9JW56 A0A2W1B072 A0A1H6FE09 A0A2S2N8L4 A0A226CY92 A0A2S2PF27 A0A3C1TPI2 D2A336 J9K8H7 J9M5W4 A0A2S2QDI3 J9L8L7 X1XDK5 A0A2H1X3K3 D7EKQ4 A0A226D544 A0A087T1E7 J9M9H5 J9M126 Q49HI7
Ontologies
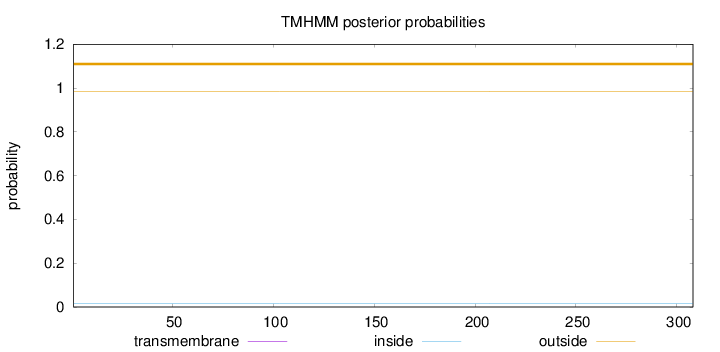
Topology
Length:
308
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00156
Exp number, first 60 AAs:
0
Total prob of N-in:
0.01471
outside
1 - 308
Population Genetic Test Statistics
Pi
4.592036
Theta
8.047277
Tajima's D
-1.587356
CLR
1.27505
CSRT
0.0435478226088696
Interpretation
Uncertain
