Gene
KWMTBOMO00500
Pre Gene Modal
BGIBMGA012310
Annotation
PREDICTED:_uncharacterized_protein_LOC105842551_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 4.935
Sequence
CDS
ATGGATACCGATAGGAAAAAAGATTTAAAAGTTAAACATAAAAAAAATCGACGTAAAAATGAAAAGAAGGAAAAAATGGAAGCAACCAATACGGATTTAAAATTGGACAATATGTGTAATCAAACCAGCAGGCTGACTGACGGTAATGGTAGTGAGTCGGGGAGCTCTACCGACAGCGAGGACTTAGTCAATGCTATAAACTACAACACCATTACGTCATTAGCAGCGCTCAAGGATATCATACAGGGTGACAATAATGATACACCAGAGACTGATAGTTTCTTCCAACATTATTTTATGAACCATTGTAATAATCTTTCAAGTAGAAATATACCTCCAAACTCACCGTTCCCATATACTGAGCGTCGACGTCTATCTCAATGCCGCGAAGAAGACGAAGAAGACGACAAAAGGGAAAACGATCCGCCTGCACCTATTTTATTTTCTGAGATAGAATTGGGAAAGCCGCATCAGTCTATTTCAGATATTTTGGCCAAAGAACTTAGCTTAAGTGATGGCAGCGAAGTAAAGCGCAGTAACGCGGAGAAAGATAAGTCACCAGATGATTTGTATGACAACGAGGATGATCACGCGCGTTTAAACCGAACTATTATGGGAGCGAAACATAAGTTTCTAGTAACAACGACAGAGCCACCCCTGGCGTCCGTTGAAAGGGCACTACGCTCTCATATACACAATGCTCACACTGTACATTTCGGGACTAAGGGTGCTGAAGAACAGAGACCGAGTATGCGATCTATTTTTACAGGACAGAATCTCCATCGTGATAAAAATTATTTTGATAGTAGCTTGATAGAAATTCGAAGTACGGTCGACGAAATCCCCACGAGCTCCGAAGATATATGGGTTAAGAGAACTGAGGATAAGACGGATGCTGCGTTAATAAAACAAACAATAATACAAGATACACCAGCTTCGATCACTAATAATGATACAGAAAAACAAAATGAAAAGCCTGAAACAAATGAAACTGAAATGCGTGATTCAAAGACGTCTGATAAGAGCACGGAGAGTGTGCGATCTCGGTCTGGTACTTGGAGTTTTCGTATGACGCCCAAAGAAAAAGATGCTTTGAATAAAATTCAATATCGTCGCGAGAAGGACTTGCGGCGTAGCAGTGATGAAAGGCGACAGAAACGTGATAACCGGCTCGATGCTGCGATCACACGCAGCTCACAGTCAGTAGGCGAACGCAAACGCAGGGGTTCTAACGATGACGGACAACCCCACCCTATGTACCAACAAACGATACATGGCACCTCCGGGGTCAGTCGCAGACCGGCATTGTTCGATATATTTAAATCGAAACCTAAAACCGGAGATGCGAAGAAACAATCATCGATCATGCAACAAGTCAAAGCTGCCGTCCAGAGTATTACCAAATCATCTGGTGGCTCTTCGACTAAGGAAGTTAAACAAGAACCTTCCAAACCGGAAGCCTCCACGAGCAGCACCACGCCAGTGGTGAAAGTGAGAGACGGATCAGCTCACAAACATGCCGGAAGCGACACAAGGTATTACCACACATTGACGGCATCGTCGTCCCGACGTCCTAGCGCCATGGCGAAGGTCATGGAAATGTTTCGAGGTCGTTCATCCGTCCCGGGTCAAGCTCCGCCAACGGATGCTGCCGAAAAAAGAAAAGCGAGGGCATTGGCTCAGCAACAACACTACGCTACAGCGGATTAG
Protein
MDTDRKKDLKVKHKKNRRKNEKKEKMEATNTDLKLDNMCNQTSRLTDGNGSESGSSTDSEDLVNAINYNTITSLAALKDIIQGDNNDTPETDSFFQHYFMNHCNNLSSRNIPPNSPFPYTERRRLSQCREEDEEDDKRENDPPAPILFSEIELGKPHQSISDILAKELSLSDGSEVKRSNAEKDKSPDDLYDNEDDHARLNRTIMGAKHKFLVTTTEPPLASVERALRSHIHNAHTVHFGTKGAEEQRPSMRSIFTGQNLHRDKNYFDSSLIEIRSTVDEIPTSSEDIWVKRTEDKTDAALIKQTIIQDTPASITNNDTEKQNEKPETNETEMRDSKTSDKSTESVRSRSGTWSFRMTPKEKDALNKIQYRREKDLRRSSDERRQKRDNRLDAAITRSSQSVGERKRRGSNDDGQPHPMYQQTIHGTSGVSRRPALFDIFKSKPKTGDAKKQSSIMQQVKAAVQSITKSSGGSSTKEVKQEPSKPEASTSSTTPVVKVRDGSAHKHAGSDTRYYHTLTASSSRRPSAMAKVMEMFRGRSSVPGQAPPTDAAEKRKARALAQQQHYATAD
Summary
Uniprot
H9JS00
A0A2H1V4F1
A0A212EIB6
A0A194PGK8
A0A1E1WM50
A0A194QU10
+ More
A0A1B1PFX8 A0A336MRL8 A0A336MA84 A0A154PE37 A0A1Y1LHQ5 A0A336MLY9 U5EQ69 A0A182NSA8 A0A182X9Z2 A0A2M4BAM7 A0A182HKX3 A0A182K0T4 A0A2M4B9X6 A0A182VYV8 A0A182UFN7 A0A182TMU6 F5HMQ3 A0A182L987 A0A182QD89 A0A182VF77 B0WG22 A0A084VCY7 A0A182ISN2 A0A1B0FLY7 A0A182MNZ3
A0A1B1PFX8 A0A336MRL8 A0A336MA84 A0A154PE37 A0A1Y1LHQ5 A0A336MLY9 U5EQ69 A0A182NSA8 A0A182X9Z2 A0A2M4BAM7 A0A182HKX3 A0A182K0T4 A0A2M4B9X6 A0A182VYV8 A0A182UFN7 A0A182TMU6 F5HMQ3 A0A182L987 A0A182QD89 A0A182VF77 B0WG22 A0A084VCY7 A0A182ISN2 A0A1B0FLY7 A0A182MNZ3
EMBL
BABH01016965
ODYU01000621
SOQ35725.1
AGBW02014676
OWR41232.1
KQ459605
+ More
KPI91844.1 GDQN01002970 JAT88084.1 KQ461108 KPJ09018.1 KX121428 ANT47201.1 UFQS01001691 UFQT01001691 SSX11870.1 SSX31433.1 UFQT01000617 SSX25799.1 KQ434886 KZC10129.1 GEZM01058142 JAV71870.1 SSX11871.1 SSX31434.1 GANO01004384 JAB55487.1 GGFJ01000964 MBW50105.1 APCN01000875 GGFJ01000679 MBW49820.1 AAAB01008987 EGK97574.1 AXCN02000206 DS231922 EDS26663.1 ATLV01011012 KE524633 KFB35831.1 CCAG010006360 CCAG010006361 CCAG010006362 CCAG010006363 CCAG010006364 CCAG010006365 CCAG010006366 AXCM01006328
KPI91844.1 GDQN01002970 JAT88084.1 KQ461108 KPJ09018.1 KX121428 ANT47201.1 UFQS01001691 UFQT01001691 SSX11870.1 SSX31433.1 UFQT01000617 SSX25799.1 KQ434886 KZC10129.1 GEZM01058142 JAV71870.1 SSX11871.1 SSX31434.1 GANO01004384 JAB55487.1 GGFJ01000964 MBW50105.1 APCN01000875 GGFJ01000679 MBW49820.1 AAAB01008987 EGK97574.1 AXCN02000206 DS231922 EDS26663.1 ATLV01011012 KE524633 KFB35831.1 CCAG010006360 CCAG010006361 CCAG010006362 CCAG010006363 CCAG010006364 CCAG010006365 CCAG010006366 AXCM01006328
Proteomes
PRIDE
Pfam
PF00571 CBS
ProteinModelPortal
H9JS00
A0A2H1V4F1
A0A212EIB6
A0A194PGK8
A0A1E1WM50
A0A194QU10
+ More
A0A1B1PFX8 A0A336MRL8 A0A336MA84 A0A154PE37 A0A1Y1LHQ5 A0A336MLY9 U5EQ69 A0A182NSA8 A0A182X9Z2 A0A2M4BAM7 A0A182HKX3 A0A182K0T4 A0A2M4B9X6 A0A182VYV8 A0A182UFN7 A0A182TMU6 F5HMQ3 A0A182L987 A0A182QD89 A0A182VF77 B0WG22 A0A084VCY7 A0A182ISN2 A0A1B0FLY7 A0A182MNZ3
A0A1B1PFX8 A0A336MRL8 A0A336MA84 A0A154PE37 A0A1Y1LHQ5 A0A336MLY9 U5EQ69 A0A182NSA8 A0A182X9Z2 A0A2M4BAM7 A0A182HKX3 A0A182K0T4 A0A2M4B9X6 A0A182VYV8 A0A182UFN7 A0A182TMU6 F5HMQ3 A0A182L987 A0A182QD89 A0A182VF77 B0WG22 A0A084VCY7 A0A182ISN2 A0A1B0FLY7 A0A182MNZ3
Ontologies
PANTHER
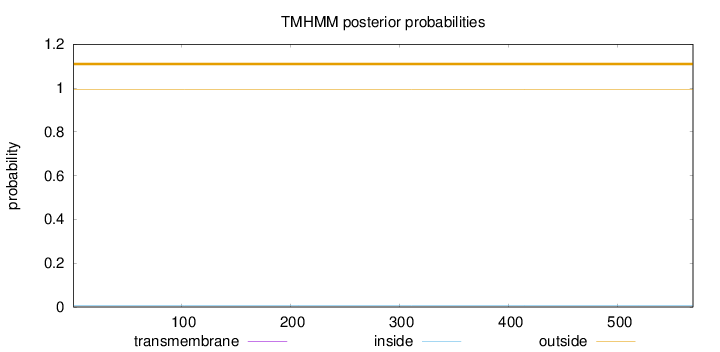
Topology
Length:
569
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00018
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00710
outside
1 - 569
Population Genetic Test Statistics
Pi
24.620526
Theta
24.126767
Tajima's D
0.007451
CLR
0.075215
CSRT
0.370831458427079
Interpretation
Uncertain
