Gene
KWMTBOMO00494
Pre Gene Modal
BGIBMGA002243
Annotation
PREDICTED:_dephospho-CoA_kinase_[Papilio_xuthus]
Location in the cell
PlasmaMembrane Reliability : 2.484
Sequence
CDS
ATGCAGTTAGTTCACGTGTTTGTGAATGTAAGGCAAGAAAGAAATATGTTCATTGTTGGACTGACTGGTGGCCTCGCAACAGGCAAGAGCACAGTGCTCTCCATATTTAAAGAACACGGAATTGCCGTAATAGATGCCGATGAAGTTGCTCGAAAAGTTTTAGAACCTGGCACAAAAGCTTGGCTTGAGGTTAAGCAGTATTTTGGACATGGAGTTCTGTTCCCTGATGGAAGAGTTAACAGATTAAAACTTGGCGAAATTGTTTTTGATGATATAGAAAAACGACGTAAACTTAATGCTATCACACATCCAAGGATACAAAGCGCAATGATAAGAATTGCATTTTCATTCTTTTTCACTGGTCATACTTATATTGTGATGGAAGTACCTCTCCTTTTTGAAACAGGAAAATTATTGACATTCATGCATAAAATTATTACTGTTGTTTGTGAGGATCACCAGCAGCTTGAACGTTTATACAAAAGGAATGACTTCTCTGAAATAGTAGCCAGAAAGAGAATTAATAGTCAAATGCCACTTCATGAAAAGGTTGCCAAATCACACTTTGTTATTGATAATTCTGGTGATCTTACAAACACCAGGAATCAGACAGAGAGTATTATTAAAGTCTTAAGAAAATCAAAGTTTACTTGGTACTTCAGAGCAATATTTATGATAACAATACTGTCAGCAGTTTTTGCTGTTACACATATTACAAGCAATTATCTTGAAAATTTATAA
Protein
MQLVHVFVNVRQERNMFIVGLTGGLATGKSTVLSIFKEHGIAVIDADEVARKVLEPGTKAWLEVKQYFGHGVLFPDGRVNRLKLGEIVFDDIEKRRKLNAITHPRIQSAMIRIAFSFFFTGHTYIVMEVPLLFETGKLLTFMHKIITVVCEDHQQLERLYKRNDFSEIVARKRINSQMPLHEKVAKSHFVIDNSGDLTNTRNQTESIIKVLRKSKFTWYFRAIFMITILSAVFAVTHITSNYLENL
Summary
Uniprot
H9IYB0
A0A2H1VAE7
S4PTB2
A0A194QZI9
A0A212EIC0
A0A2A4J1G3
+ More
A0A194PL65 A0A2P8XPM7 A0A2J7QG42 A0A067QLL2 A0A336KM79 A0A182H451 A0A1B6EDM8 A0A151IJT1 A0A2A3EB86 E9J7U2 A0A151WRD8 A0A1B0D2B6 A0A2R7VZ33 A0A151JB55 A0A1L8DZF8 Q16JB1 E2BTT0 A0A195FC49 A0A154NWD6 A0A1B6FZB3 A0A158NKI7 A0A195B080 A0A0M4EUB5 A0A088A2G9 A0A0L7RG40 A0A1I8M209 A0A1Q3G362 E2A3Q7 A0A131YM05 A0A310SMY6 A0A1B0CAM8 A0A182JIE8 A0A210PFV5 A0A131XED4 U5EZN6 Q8SWY7 A0A0M8ZS45 T1DHG1 B4LZC7 B4KDA2 A0A0P4XA34 B4JHW2 A0A2M4BYK3 L7ME38 L7MBB5 Q293K8 A0A2M3ZB03 B4GLZ9 B4QYZ6 Q9VI57 A0A2M4ARA8 B4I4N4 B4PS21 B3LYC3 A0A1I8PIJ1 A0A3B0JPX4 B3P2S5 A0A2C9H8U9 A0A182UQV3 A0A182UEH3 A0A182KZS3 Q7QA58 A0A182HVQ2 A0A182Y8Z4 A0A182W397 W5JWR0 A0A1W4UEY6 G3MQJ8 A0A023GAS7 A0A182RI12 A0A0L0BZP5 A0A182S5E8 F4WWB4 A0A182QHU2 A0A0K8VJ44 A0A0K8UPD9 T1JD75 A0A182NCW3 A0A293LM07 A0A1A9WLU4 B4NKM2 A0A034WUH9 A0A131XXV5 V5IHB7 A0A0A1XCS6 A0A0A9Y3C7 A0A087TIL9 A0A084WPA5 A0A226DWX4 A0A0K8SW49 A0A069DQY4 A0A2R5LHJ7 A0A2P6L6Z6
A0A194PL65 A0A2P8XPM7 A0A2J7QG42 A0A067QLL2 A0A336KM79 A0A182H451 A0A1B6EDM8 A0A151IJT1 A0A2A3EB86 E9J7U2 A0A151WRD8 A0A1B0D2B6 A0A2R7VZ33 A0A151JB55 A0A1L8DZF8 Q16JB1 E2BTT0 A0A195FC49 A0A154NWD6 A0A1B6FZB3 A0A158NKI7 A0A195B080 A0A0M4EUB5 A0A088A2G9 A0A0L7RG40 A0A1I8M209 A0A1Q3G362 E2A3Q7 A0A131YM05 A0A310SMY6 A0A1B0CAM8 A0A182JIE8 A0A210PFV5 A0A131XED4 U5EZN6 Q8SWY7 A0A0M8ZS45 T1DHG1 B4LZC7 B4KDA2 A0A0P4XA34 B4JHW2 A0A2M4BYK3 L7ME38 L7MBB5 Q293K8 A0A2M3ZB03 B4GLZ9 B4QYZ6 Q9VI57 A0A2M4ARA8 B4I4N4 B4PS21 B3LYC3 A0A1I8PIJ1 A0A3B0JPX4 B3P2S5 A0A2C9H8U9 A0A182UQV3 A0A182UEH3 A0A182KZS3 Q7QA58 A0A182HVQ2 A0A182Y8Z4 A0A182W397 W5JWR0 A0A1W4UEY6 G3MQJ8 A0A023GAS7 A0A182RI12 A0A0L0BZP5 A0A182S5E8 F4WWB4 A0A182QHU2 A0A0K8VJ44 A0A0K8UPD9 T1JD75 A0A182NCW3 A0A293LM07 A0A1A9WLU4 B4NKM2 A0A034WUH9 A0A131XXV5 V5IHB7 A0A0A1XCS6 A0A0A9Y3C7 A0A087TIL9 A0A084WPA5 A0A226DWX4 A0A0K8SW49 A0A069DQY4 A0A2R5LHJ7 A0A2P6L6Z6
Pubmed
19121390
23622113
26354079
22118469
29403074
24845553
+ More
26483478 21282665 17510324 20798317 21347285 25315136 26830274 28812685 28049606 17994087 25576852 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20966253 12364791 14747013 17210077 25244985 20920257 23761445 22216098 26108605 21719571 25348373 25765539 25830018 25401762 26823975 24438588 26334808
26483478 21282665 17510324 20798317 21347285 25315136 26830274 28812685 28049606 17994087 25576852 15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 17550304 20966253 12364791 14747013 17210077 25244985 20920257 23761445 22216098 26108605 21719571 25348373 25765539 25830018 25401762 26823975 24438588 26334808
EMBL
BABH01020862
ODYU01001491
SOQ37771.1
GAIX01012343
JAA80217.1
KQ461108
+ More
KPJ09011.1 AGBW02014676 OWR41224.1 NWSH01004009 PCG65576.1 KQ459605 KPI91850.1 PYGN01001573 PSN33962.1 NEVH01014835 PNF27564.1 KK853191 KDR10023.1 UFQS01000590 UFQT01000590 SSX05172.1 SSX25533.1 JXUM01005321 JXUM01005322 JXUM01108788 JXUM01108789 KQ565264 KQ560191 KXJ71162.1 KXJ83901.1 GEDC01026663 GEDC01006771 GEDC01001291 JAS10635.1 JAS30527.1 JAS36007.1 KQ977294 KYN03909.1 KZ288311 PBC28469.1 GL768630 EFZ11119.1 KQ982805 KYQ50469.1 AJVK01022730 KK854175 PTY12599.1 KQ979182 KYN22337.1 GFDF01002277 JAV11807.1 CH478020 EAT34348.1 GL450531 EFN80829.1 KQ981693 KYN37777.1 KQ434772 KZC03903.1 GECZ01014245 JAS55524.1 ADTU01018855 KQ976692 KYM77866.1 CP012526 ALC46655.1 KQ414601 KOC69701.1 GFDL01000787 JAV34258.1 GL436457 EFN71949.1 GEDV01009626 JAP78931.1 KQ759843 OAD62605.1 AJWK01004116 NEDP02076731 OWF35365.1 GEFH01002647 JAP65934.1 GANO01000454 JAB59417.1 AY094946 AAM11299.1 KQ435881 KOX69837.1 GAMD01002376 JAA99214.1 CH940650 EDW68162.1 CH933806 EDW14884.1 GDIP01244763 JAI78638.1 CH916369 EDV92870.1 GGFJ01008951 MBW58092.1 GACK01002543 JAA62491.1 GACK01003897 JAA61137.1 CM000070 EAL29207.2 GGFM01004904 MBW25655.1 CH479185 EDW38573.1 CM000364 EDX11936.1 AE014297 BT128902 AAF54069.1 AEO51075.1 GGFK01009931 MBW43252.1 CH480821 EDW55177.1 CM000160 EDW96421.1 CH902617 EDV41786.1 OUUW01000008 SPP84175.1 CH954181 EDV48097.1 AAAB01008898 EAA09265.3 APCN01005809 ADMH02000180 ETN67474.1 JO844149 AEO35766.1 GBBM01005415 JAC30003.1 JRES01001097 KNC25547.1 GL888406 EGI61456.1 AXCN02001559 GDHF01013405 JAI38909.1 GDHF01023750 JAI28564.1 JH432100 GFWV01004366 MAA29096.1 CH964272 EDW85194.1 GAKP01001097 JAC57855.1 GEFM01004751 JAP71045.1 GANP01005217 JAB79251.1 GBXI01005547 JAD08745.1 GBHO01016935 GDHC01018319 JAG26669.1 JAQ00310.1 KK115375 KFM64958.1 ATLV01025001 ATLV01025002 ATLV01025003 ATLV01025004 KE525367 KFB52049.1 LNIX01000009 OXA49972.1 GBRD01008711 JAG57110.1 GBGD01002768 JAC86121.1 GGLE01004833 MBY08959.1 MWRG01001338 PRD34370.1
KPJ09011.1 AGBW02014676 OWR41224.1 NWSH01004009 PCG65576.1 KQ459605 KPI91850.1 PYGN01001573 PSN33962.1 NEVH01014835 PNF27564.1 KK853191 KDR10023.1 UFQS01000590 UFQT01000590 SSX05172.1 SSX25533.1 JXUM01005321 JXUM01005322 JXUM01108788 JXUM01108789 KQ565264 KQ560191 KXJ71162.1 KXJ83901.1 GEDC01026663 GEDC01006771 GEDC01001291 JAS10635.1 JAS30527.1 JAS36007.1 KQ977294 KYN03909.1 KZ288311 PBC28469.1 GL768630 EFZ11119.1 KQ982805 KYQ50469.1 AJVK01022730 KK854175 PTY12599.1 KQ979182 KYN22337.1 GFDF01002277 JAV11807.1 CH478020 EAT34348.1 GL450531 EFN80829.1 KQ981693 KYN37777.1 KQ434772 KZC03903.1 GECZ01014245 JAS55524.1 ADTU01018855 KQ976692 KYM77866.1 CP012526 ALC46655.1 KQ414601 KOC69701.1 GFDL01000787 JAV34258.1 GL436457 EFN71949.1 GEDV01009626 JAP78931.1 KQ759843 OAD62605.1 AJWK01004116 NEDP02076731 OWF35365.1 GEFH01002647 JAP65934.1 GANO01000454 JAB59417.1 AY094946 AAM11299.1 KQ435881 KOX69837.1 GAMD01002376 JAA99214.1 CH940650 EDW68162.1 CH933806 EDW14884.1 GDIP01244763 JAI78638.1 CH916369 EDV92870.1 GGFJ01008951 MBW58092.1 GACK01002543 JAA62491.1 GACK01003897 JAA61137.1 CM000070 EAL29207.2 GGFM01004904 MBW25655.1 CH479185 EDW38573.1 CM000364 EDX11936.1 AE014297 BT128902 AAF54069.1 AEO51075.1 GGFK01009931 MBW43252.1 CH480821 EDW55177.1 CM000160 EDW96421.1 CH902617 EDV41786.1 OUUW01000008 SPP84175.1 CH954181 EDV48097.1 AAAB01008898 EAA09265.3 APCN01005809 ADMH02000180 ETN67474.1 JO844149 AEO35766.1 GBBM01005415 JAC30003.1 JRES01001097 KNC25547.1 GL888406 EGI61456.1 AXCN02001559 GDHF01013405 JAI38909.1 GDHF01023750 JAI28564.1 JH432100 GFWV01004366 MAA29096.1 CH964272 EDW85194.1 GAKP01001097 JAC57855.1 GEFM01004751 JAP71045.1 GANP01005217 JAB79251.1 GBXI01005547 JAD08745.1 GBHO01016935 GDHC01018319 JAG26669.1 JAQ00310.1 KK115375 KFM64958.1 ATLV01025001 ATLV01025002 ATLV01025003 ATLV01025004 KE525367 KFB52049.1 LNIX01000009 OXA49972.1 GBRD01008711 JAG57110.1 GBGD01002768 JAC86121.1 GGLE01004833 MBY08959.1 MWRG01001338 PRD34370.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000218220
UP000053268
UP000245037
+ More
UP000235965 UP000027135 UP000069940 UP000249989 UP000078542 UP000242457 UP000075809 UP000092462 UP000078492 UP000008820 UP000008237 UP000078541 UP000076502 UP000005205 UP000078540 UP000092553 UP000005203 UP000053825 UP000095301 UP000000311 UP000092461 UP000075880 UP000242188 UP000053105 UP000008792 UP000009192 UP000001070 UP000001819 UP000008744 UP000000304 UP000000803 UP000001292 UP000002282 UP000007801 UP000095300 UP000268350 UP000008711 UP000076407 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840 UP000076408 UP000075920 UP000000673 UP000192221 UP000075900 UP000037069 UP000075901 UP000007755 UP000075886 UP000075884 UP000091820 UP000007798 UP000054359 UP000030765 UP000198287
UP000235965 UP000027135 UP000069940 UP000249989 UP000078542 UP000242457 UP000075809 UP000092462 UP000078492 UP000008820 UP000008237 UP000078541 UP000076502 UP000005205 UP000078540 UP000092553 UP000005203 UP000053825 UP000095301 UP000000311 UP000092461 UP000075880 UP000242188 UP000053105 UP000008792 UP000009192 UP000001070 UP000001819 UP000008744 UP000000304 UP000000803 UP000001292 UP000002282 UP000007801 UP000095300 UP000268350 UP000008711 UP000076407 UP000075903 UP000075902 UP000075882 UP000007062 UP000075840 UP000076408 UP000075920 UP000000673 UP000192221 UP000075900 UP000037069 UP000075901 UP000007755 UP000075886 UP000075884 UP000091820 UP000007798 UP000054359 UP000030765 UP000198287
Pfam
PF01121 CoaE
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
H9IYB0
A0A2H1VAE7
S4PTB2
A0A194QZI9
A0A212EIC0
A0A2A4J1G3
+ More
A0A194PL65 A0A2P8XPM7 A0A2J7QG42 A0A067QLL2 A0A336KM79 A0A182H451 A0A1B6EDM8 A0A151IJT1 A0A2A3EB86 E9J7U2 A0A151WRD8 A0A1B0D2B6 A0A2R7VZ33 A0A151JB55 A0A1L8DZF8 Q16JB1 E2BTT0 A0A195FC49 A0A154NWD6 A0A1B6FZB3 A0A158NKI7 A0A195B080 A0A0M4EUB5 A0A088A2G9 A0A0L7RG40 A0A1I8M209 A0A1Q3G362 E2A3Q7 A0A131YM05 A0A310SMY6 A0A1B0CAM8 A0A182JIE8 A0A210PFV5 A0A131XED4 U5EZN6 Q8SWY7 A0A0M8ZS45 T1DHG1 B4LZC7 B4KDA2 A0A0P4XA34 B4JHW2 A0A2M4BYK3 L7ME38 L7MBB5 Q293K8 A0A2M3ZB03 B4GLZ9 B4QYZ6 Q9VI57 A0A2M4ARA8 B4I4N4 B4PS21 B3LYC3 A0A1I8PIJ1 A0A3B0JPX4 B3P2S5 A0A2C9H8U9 A0A182UQV3 A0A182UEH3 A0A182KZS3 Q7QA58 A0A182HVQ2 A0A182Y8Z4 A0A182W397 W5JWR0 A0A1W4UEY6 G3MQJ8 A0A023GAS7 A0A182RI12 A0A0L0BZP5 A0A182S5E8 F4WWB4 A0A182QHU2 A0A0K8VJ44 A0A0K8UPD9 T1JD75 A0A182NCW3 A0A293LM07 A0A1A9WLU4 B4NKM2 A0A034WUH9 A0A131XXV5 V5IHB7 A0A0A1XCS6 A0A0A9Y3C7 A0A087TIL9 A0A084WPA5 A0A226DWX4 A0A0K8SW49 A0A069DQY4 A0A2R5LHJ7 A0A2P6L6Z6
A0A194PL65 A0A2P8XPM7 A0A2J7QG42 A0A067QLL2 A0A336KM79 A0A182H451 A0A1B6EDM8 A0A151IJT1 A0A2A3EB86 E9J7U2 A0A151WRD8 A0A1B0D2B6 A0A2R7VZ33 A0A151JB55 A0A1L8DZF8 Q16JB1 E2BTT0 A0A195FC49 A0A154NWD6 A0A1B6FZB3 A0A158NKI7 A0A195B080 A0A0M4EUB5 A0A088A2G9 A0A0L7RG40 A0A1I8M209 A0A1Q3G362 E2A3Q7 A0A131YM05 A0A310SMY6 A0A1B0CAM8 A0A182JIE8 A0A210PFV5 A0A131XED4 U5EZN6 Q8SWY7 A0A0M8ZS45 T1DHG1 B4LZC7 B4KDA2 A0A0P4XA34 B4JHW2 A0A2M4BYK3 L7ME38 L7MBB5 Q293K8 A0A2M3ZB03 B4GLZ9 B4QYZ6 Q9VI57 A0A2M4ARA8 B4I4N4 B4PS21 B3LYC3 A0A1I8PIJ1 A0A3B0JPX4 B3P2S5 A0A2C9H8U9 A0A182UQV3 A0A182UEH3 A0A182KZS3 Q7QA58 A0A182HVQ2 A0A182Y8Z4 A0A182W397 W5JWR0 A0A1W4UEY6 G3MQJ8 A0A023GAS7 A0A182RI12 A0A0L0BZP5 A0A182S5E8 F4WWB4 A0A182QHU2 A0A0K8VJ44 A0A0K8UPD9 T1JD75 A0A182NCW3 A0A293LM07 A0A1A9WLU4 B4NKM2 A0A034WUH9 A0A131XXV5 V5IHB7 A0A0A1XCS6 A0A0A9Y3C7 A0A087TIL9 A0A084WPA5 A0A226DWX4 A0A0K8SW49 A0A069DQY4 A0A2R5LHJ7 A0A2P6L6Z6
PDB
2IF2
E-value=4.70223e-24,
Score=273
Ontologies
GO
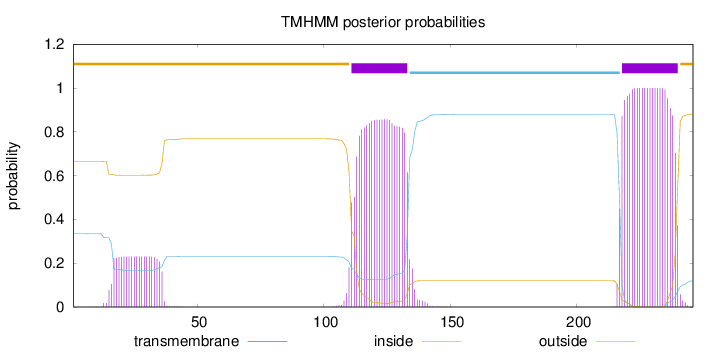
Topology
Length:
246
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
46.02174
Exp number, first 60 AAs:
4.75003
Total prob of N-in:
0.33429
outside
1 - 110
TMhelix
111 - 133
inside
134 - 217
TMhelix
218 - 240
outside
241 - 246
Population Genetic Test Statistics
Pi
14.830714
Theta
13.282852
Tajima's D
0.344656
CLR
0
CSRT
0.47997600119994
Interpretation
Uncertain
