Gene
KWMTBOMO00493 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA002337
Annotation
PREDICTED:_enolase_[Papilio_xuthus]
Transcription factor
Location in the cell
Mitochondrial Reliability : 0.769
Sequence
CDS
ATGTTCGATATGGACGGTGTGAGCGGCATCAGATGCCAAGTGATTGACAATAGCAGTGCTGTGCCCGTCGCCGTAGGTGGATCTCCGACATCCGCAAATGGATCCGACGTGTTCTTATACGACGACTCCTCAATGTCCGACGGCAGCCTATATGCTAGTGATCAGGAAAATCATTCTACGCCAAGAAGAAGATCAGAATACCGGCGCGTTCACAAGCGACGCTCGCGTTGTCCGCAGCAACAAATCCAACAGAGGCAAGCCGCGAATCTTCGGGAAAGACGTCGAATGCAATCCATTAATGACGCTTTTGAAGGTTTAAGAGCACACATTCCAACATTGCCTTATGAGAAACGTCTGTCCAAAGTCGATACCTTGAAGCTTGCGATCGGATACATCAGTTTTCTTGGAGAATTAGTACGGGCTGATCGCAATTCATCCGACGGGTTGTTGTCAGGTGTGGGCAACACTCAGAATAAAGAAGAACAAAAGAAAATCATTATTAGAGAACCTTTATTCATTTATAAAGTCAAGATGCCAATTAAGTTATTGCTAGCACGTCAAATTTTTGACTCCACTGGTGTCCCTACAGTCGAAGTAGATATGGTGACAGAATTAGGTCTATTTCGAATAGGTGTACCTTCCACGGACTCCAAAAAAATAGCCGAAGCAACACAACTGCGGGATAACAATCCTGCTCAATACTTTGGAATGGGTGTTTCTAGTGCTATCAAAAATATTAACGTCATTATTGCACCTGAACTTATCAAACAAAACCTTGAAGTGACTATGCAAAAGGAAATTGATCAATTCTTGATATCTCTCGATGGAACTGAAAACCGATCTAGACTAGGAGCTAACGCGATTCTTTGTGTTTCTCTGGTAGTCGCAAAAGCTGGAGCTGCGAAGAAAGGTGTACCTTTGTATAGGCACATATCGGATATGGCAGGCGTCACTACAATAATTCTGCCAGTACCACACTTTACTATTCTCACTGGTGGAATTTTATCTTCAAATGGACTGCCGTTTCAAGAATACATAATTATGCCTACAGGAGCATCTTCGTTTGCAGATGCGATGCGTATTGGTTCAGAAATTTATCGTTATGTGAAAAACGCGATTGCGAATAAGTTTTCTGTAGACTGCACGTATGTTAGTGATTCCGGAGGATTTTCTGTGCCCTTGCAAAGTCATCGGGACGCTCTTATGTTTCTTACAGATGCTATTAAGCAATGTGGGTATGTTGGCAAAGCTGAAATTGCAATAAATGCTGCCGCCAGTGACATGTTCAAAGATGGTGCCTACGATTTGGAGTTTAAAAATCCGAACTCAAATCCTCAGGAATACATGTCGTCGGATAAACTTGCCGAAATTTATCTAGATAATATAAAAGAGTTTCCAGTATGTTCTATTGAAGACGGTTTCCAATTTGATGATTGGTCGGCGTGGTCTACTTTGACCTCTCGAACGCAGGCTCAGATTTTTGGAAATGACCTTACTCAAACTAATCTCCGCCGTGTTGGATTGGCCACAGAAAAAAAAGCGGGAAATGCGATTGGCTTAAGACTAAATCAAGCTGGCACGTTTACCGAAGTTATTGAAGCTTTTAAACTTTTGAAGTCAAATGGTTTTGCTTCTGTTGTTGTTGATAGGTGGGGTGACACTGAAGATGTGTTCCTAGCCGATCTGGCGGTTGGTTTGTCTACTGGTCAGATGAAATGCGGAGCACCTGTTCGATCTGAACGCGTCGGAAAATACAATCAAATCATTAGAATTGAAGAAGAATTAGGGGCGCTTTCCAAATACGCCGGAAAAAACTACCGCGGACTAAATTAA
Protein
MFDMDGVSGIRCQVIDNSSAVPVAVGGSPTSANGSDVFLYDDSSMSDGSLYASDQENHSTPRRRSEYRRVHKRRSRCPQQQIQQRQAANLRERRRMQSINDAFEGLRAHIPTLPYEKRLSKVDTLKLAIGYISFLGELVRADRNSSDGLLSGVGNTQNKEEQKKIIIREPLFIYKVKMPIKLLLARQIFDSTGVPTVEVDMVTELGLFRIGVPSTDSKKIAEATQLRDNNPAQYFGMGVSSAIKNINVIIAPELIKQNLEVTMQKEIDQFLISLDGTENRSRLGANAILCVSLVVAKAGAAKKGVPLYRHISDMAGVTTIILPVPHFTILTGGILSSNGLPFQEYIIMPTGASSFADAMRIGSEIYRYVKNAIANKFSVDCTYVSDSGGFSVPLQSHRDALMFLTDAIKQCGYVGKAEIAINAAASDMFKDGAYDLEFKNPNSNPQEYMSSDKLAEIYLDNIKEFPVCSIEDGFQFDDWSAWSTLTSRTQAQIFGNDLTQTNLRRVGLATEKKAGNAIGLRLNQAGTFTEVIEAFKLLKSNGFASVVVDRWGDTEDVFLADLAVGLSTGQMKCGAPVRSERVGKYNQIIRIEEELGALSKYAGKNYRGLN
Summary
Uniprot
A0A2H1VA92
A0A212EI84
A0A0L7KWK3
H9IYK3
A0A1L7P086
E0AE73
+ More
A0A194QUL0 A0A194QY73 I4DJA8 S5G1I9 S4PLS1 A0A0K2CU86 E3T796 F0UYI6 H9J7K1 A0A1L7P084 A1YQ87 A0A0L7LLA9 A0A1D1YYT3 V5SHY9 A0A1W4WXQ7 D6X009 A0A0C9RZV9 A0A034VN28 A0A0K8VLF5 A0A0T6B9W0 A0A1L8E1V7 W8BG43 A0A067RMN9 A0A1L8E1X5 R4V1A4 A0A0L0CCJ1 A0A0A1WYC5 W8GMW5 J3JXB7 A0A1Y1KVP6 A0A1A9WZY7 T1PF13 A0A1A9Y778 A0A1L8EAC8 A0A1L8EAL7 A0A1L8EAH8 A0A1B0BRJ2 A0A0K8TU66 A0A2J7QE60 A0A1I8NVJ6 A8CWB5 A0A1I8NCM7 A0A1B0AJN7 A0A1L8EAH5 A0A182P0G3 D3TNV2 A0A1A9UMG6 A0A1Y9H9P7 A0A1B0FIL0 A0A240PKU2 A0A087SYD4 A0A182SGM0 A0A2C9GUE1 E9GAM4 A0A1S4GX21 A0A1C7CZ11 A0A182UQ73 A0A182U3K2 A0A182X1L6 A0A182IEJ8 A0A182QRK1 A0A0P5ZHM8 A0A240PJZ3 Q2HXW9 A0A182MR29 A0A182XZJ5 A0A2C9H419 A0A1Y9HCG0 A0A1I8JVF3 A0A1I8JV65 A0A182L2F0 A0A1Y9IM13 A0A1I8JT09 Q7Q3D8 A0A182MZE9 T1E772 A0A2M4A0M5 A0A2M4BNJ2 A0A182JPU4 A0A2P0XIF0 A0A1I8JSH0 A0A1Y9IVW2 A0A0P5CXY1 G9C5D8 A0A2L2Y0Q4 A0A182RVL0 A0A1B6GJX3 U5EVU9 A0A151IXY3 A0A0P5T616 T1DC53 A0A0N8CXU9 A0A2M3YZD2 A0A0P5SG68
A0A194QUL0 A0A194QY73 I4DJA8 S5G1I9 S4PLS1 A0A0K2CU86 E3T796 F0UYI6 H9J7K1 A0A1L7P084 A1YQ87 A0A0L7LLA9 A0A1D1YYT3 V5SHY9 A0A1W4WXQ7 D6X009 A0A0C9RZV9 A0A034VN28 A0A0K8VLF5 A0A0T6B9W0 A0A1L8E1V7 W8BG43 A0A067RMN9 A0A1L8E1X5 R4V1A4 A0A0L0CCJ1 A0A0A1WYC5 W8GMW5 J3JXB7 A0A1Y1KVP6 A0A1A9WZY7 T1PF13 A0A1A9Y778 A0A1L8EAC8 A0A1L8EAL7 A0A1L8EAH8 A0A1B0BRJ2 A0A0K8TU66 A0A2J7QE60 A0A1I8NVJ6 A8CWB5 A0A1I8NCM7 A0A1B0AJN7 A0A1L8EAH5 A0A182P0G3 D3TNV2 A0A1A9UMG6 A0A1Y9H9P7 A0A1B0FIL0 A0A240PKU2 A0A087SYD4 A0A182SGM0 A0A2C9GUE1 E9GAM4 A0A1S4GX21 A0A1C7CZ11 A0A182UQ73 A0A182U3K2 A0A182X1L6 A0A182IEJ8 A0A182QRK1 A0A0P5ZHM8 A0A240PJZ3 Q2HXW9 A0A182MR29 A0A182XZJ5 A0A2C9H419 A0A1Y9HCG0 A0A1I8JVF3 A0A1I8JV65 A0A182L2F0 A0A1Y9IM13 A0A1I8JT09 Q7Q3D8 A0A182MZE9 T1E772 A0A2M4A0M5 A0A2M4BNJ2 A0A182JPU4 A0A2P0XIF0 A0A1I8JSH0 A0A1Y9IVW2 A0A0P5CXY1 G9C5D8 A0A2L2Y0Q4 A0A182RVL0 A0A1B6GJX3 U5EVU9 A0A151IXY3 A0A0P5T616 T1DC53 A0A0N8CXU9 A0A2M3YZD2 A0A0P5SG68
Pubmed
EMBL
ODYU01001491
SOQ37770.1
AGBW02014676
OWR41223.1
JTDY01004989
KOB67500.1
+ More
BABH01020861 LC170037 BAW35595.1 HM755879 ADK98422.1 KQ461108 KPJ09009.1 KQ461154 KPJ08526.1 AK401376 KQ459595 BAM17998.1 KPI95597.1 KC832292 AGQ53952.1 GAIX01003885 JAA88675.1 KT218671 ALA09391.1 GU289926 ADO40102.1 FJ882068 ACZ68117.1 BABH01013947 BABH01013948 BABH01013949 LC170036 BAW35594.1 EF121427 ABL73887.1 JTDY01000648 KOB76343.1 GDJX01008128 JAT59808.1 KF647623 AHB50485.1 KQ971372 EFA10506.1 GBYB01013676 JAG83443.1 GAKP01015979 JAC42973.1 GDHF01012929 JAI39385.1 LJIG01009043 KRT83869.1 GFDF01001364 JAV12720.1 GAMC01010637 JAB95918.1 KK852575 KDR20985.1 GFDF01001363 JAV12721.1 KC571898 AGM32397.1 JRES01000569 KNC30153.1 GBXI01010213 JAD04079.1 KF986614 AHK26792.1 APGK01052696 APGK01052697 BT127886 KB741213 KB631874 AEE62848.1 ENN72673.1 ERL86865.1 GEZM01072559 GEZM01072558 JAV65469.1 KA647336 AFP61965.1 GFDG01003185 JAV15614.1 GFDG01003186 JAV15613.1 GFDG01003187 JAV15612.1 JXJN01019124 GDAI01000113 JAI17490.1 NEVH01015327 PNF26872.1 AJWK01000758 EU124610 ABV60328.1 GFDG01003183 JAV15616.1 EZ423104 ADD19380.1 AXCN02000606 CCAG010016357 KK112521 KFM57873.1 AXCM01006351 GL732537 EFX83276.1 AAAB01008964 APCN01005204 GDIP01043569 JAM60146.1 DQ368397 ABC96322.1 EAA12254.2 GAMD01003302 JAA98288.1 GGFK01000988 MBW34309.1 GGFJ01005506 MBW54647.1 KY661307 AVA17394.1 GDIP01165135 GDIP01037855 JAJ58267.1 HQ851379 AEV89757.1 IAAA01000238 IAAA01000239 IAAA01000240 LAA01779.1 GECZ01007051 JAS62718.1 GANO01003248 JAB56623.1 KQ980796 KYN12966.1 GDIP01132804 JAL70910.1 GAKT01000142 JAA92920.1 GDIP01088255 JAM15460.1 GGFM01000879 MBW21630.1 GDIP01140149 JAL63565.1
BABH01020861 LC170037 BAW35595.1 HM755879 ADK98422.1 KQ461108 KPJ09009.1 KQ461154 KPJ08526.1 AK401376 KQ459595 BAM17998.1 KPI95597.1 KC832292 AGQ53952.1 GAIX01003885 JAA88675.1 KT218671 ALA09391.1 GU289926 ADO40102.1 FJ882068 ACZ68117.1 BABH01013947 BABH01013948 BABH01013949 LC170036 BAW35594.1 EF121427 ABL73887.1 JTDY01000648 KOB76343.1 GDJX01008128 JAT59808.1 KF647623 AHB50485.1 KQ971372 EFA10506.1 GBYB01013676 JAG83443.1 GAKP01015979 JAC42973.1 GDHF01012929 JAI39385.1 LJIG01009043 KRT83869.1 GFDF01001364 JAV12720.1 GAMC01010637 JAB95918.1 KK852575 KDR20985.1 GFDF01001363 JAV12721.1 KC571898 AGM32397.1 JRES01000569 KNC30153.1 GBXI01010213 JAD04079.1 KF986614 AHK26792.1 APGK01052696 APGK01052697 BT127886 KB741213 KB631874 AEE62848.1 ENN72673.1 ERL86865.1 GEZM01072559 GEZM01072558 JAV65469.1 KA647336 AFP61965.1 GFDG01003185 JAV15614.1 GFDG01003186 JAV15613.1 GFDG01003187 JAV15612.1 JXJN01019124 GDAI01000113 JAI17490.1 NEVH01015327 PNF26872.1 AJWK01000758 EU124610 ABV60328.1 GFDG01003183 JAV15616.1 EZ423104 ADD19380.1 AXCN02000606 CCAG010016357 KK112521 KFM57873.1 AXCM01006351 GL732537 EFX83276.1 AAAB01008964 APCN01005204 GDIP01043569 JAM60146.1 DQ368397 ABC96322.1 EAA12254.2 GAMD01003302 JAA98288.1 GGFK01000988 MBW34309.1 GGFJ01005506 MBW54647.1 KY661307 AVA17394.1 GDIP01165135 GDIP01037855 JAJ58267.1 HQ851379 AEV89757.1 IAAA01000238 IAAA01000239 IAAA01000240 LAA01779.1 GECZ01007051 JAS62718.1 GANO01003248 JAB56623.1 KQ980796 KYN12966.1 GDIP01132804 JAL70910.1 GAKT01000142 JAA92920.1 GDIP01088255 JAM15460.1 GGFM01000879 MBW21630.1 GDIP01140149 JAL63565.1
Proteomes
UP000007151
UP000037510
UP000005204
UP000053240
UP000053268
UP000192223
+ More
UP000007266 UP000027135 UP000037069 UP000019118 UP000030742 UP000091820 UP000092443 UP000092460 UP000235965 UP000095300 UP000092461 UP000095301 UP000092445 UP000075885 UP000078200 UP000075886 UP000092444 UP000054359 UP000075901 UP000075883 UP000000305 UP000075882 UP000075903 UP000075902 UP000076407 UP000075840 UP000075881 UP000076408 UP000075900 UP000007062 UP000075884 UP000069272 UP000075920 UP000078492
UP000007266 UP000027135 UP000037069 UP000019118 UP000030742 UP000091820 UP000092443 UP000092460 UP000235965 UP000095300 UP000092461 UP000095301 UP000092445 UP000075885 UP000078200 UP000075886 UP000092444 UP000054359 UP000075901 UP000075883 UP000000305 UP000075882 UP000075903 UP000075902 UP000076407 UP000075840 UP000075881 UP000076408 UP000075900 UP000007062 UP000075884 UP000069272 UP000075920 UP000078492
Interpro
Gene 3D
ProteinModelPortal
A0A2H1VA92
A0A212EI84
A0A0L7KWK3
H9IYK3
A0A1L7P086
E0AE73
+ More
A0A194QUL0 A0A194QY73 I4DJA8 S5G1I9 S4PLS1 A0A0K2CU86 E3T796 F0UYI6 H9J7K1 A0A1L7P084 A1YQ87 A0A0L7LLA9 A0A1D1YYT3 V5SHY9 A0A1W4WXQ7 D6X009 A0A0C9RZV9 A0A034VN28 A0A0K8VLF5 A0A0T6B9W0 A0A1L8E1V7 W8BG43 A0A067RMN9 A0A1L8E1X5 R4V1A4 A0A0L0CCJ1 A0A0A1WYC5 W8GMW5 J3JXB7 A0A1Y1KVP6 A0A1A9WZY7 T1PF13 A0A1A9Y778 A0A1L8EAC8 A0A1L8EAL7 A0A1L8EAH8 A0A1B0BRJ2 A0A0K8TU66 A0A2J7QE60 A0A1I8NVJ6 A8CWB5 A0A1I8NCM7 A0A1B0AJN7 A0A1L8EAH5 A0A182P0G3 D3TNV2 A0A1A9UMG6 A0A1Y9H9P7 A0A1B0FIL0 A0A240PKU2 A0A087SYD4 A0A182SGM0 A0A2C9GUE1 E9GAM4 A0A1S4GX21 A0A1C7CZ11 A0A182UQ73 A0A182U3K2 A0A182X1L6 A0A182IEJ8 A0A182QRK1 A0A0P5ZHM8 A0A240PJZ3 Q2HXW9 A0A182MR29 A0A182XZJ5 A0A2C9H419 A0A1Y9HCG0 A0A1I8JVF3 A0A1I8JV65 A0A182L2F0 A0A1Y9IM13 A0A1I8JT09 Q7Q3D8 A0A182MZE9 T1E772 A0A2M4A0M5 A0A2M4BNJ2 A0A182JPU4 A0A2P0XIF0 A0A1I8JSH0 A0A1Y9IVW2 A0A0P5CXY1 G9C5D8 A0A2L2Y0Q4 A0A182RVL0 A0A1B6GJX3 U5EVU9 A0A151IXY3 A0A0P5T616 T1DC53 A0A0N8CXU9 A0A2M3YZD2 A0A0P5SG68
A0A194QUL0 A0A194QY73 I4DJA8 S5G1I9 S4PLS1 A0A0K2CU86 E3T796 F0UYI6 H9J7K1 A0A1L7P084 A1YQ87 A0A0L7LLA9 A0A1D1YYT3 V5SHY9 A0A1W4WXQ7 D6X009 A0A0C9RZV9 A0A034VN28 A0A0K8VLF5 A0A0T6B9W0 A0A1L8E1V7 W8BG43 A0A067RMN9 A0A1L8E1X5 R4V1A4 A0A0L0CCJ1 A0A0A1WYC5 W8GMW5 J3JXB7 A0A1Y1KVP6 A0A1A9WZY7 T1PF13 A0A1A9Y778 A0A1L8EAC8 A0A1L8EAL7 A0A1L8EAH8 A0A1B0BRJ2 A0A0K8TU66 A0A2J7QE60 A0A1I8NVJ6 A8CWB5 A0A1I8NCM7 A0A1B0AJN7 A0A1L8EAH5 A0A182P0G3 D3TNV2 A0A1A9UMG6 A0A1Y9H9P7 A0A1B0FIL0 A0A240PKU2 A0A087SYD4 A0A182SGM0 A0A2C9GUE1 E9GAM4 A0A1S4GX21 A0A1C7CZ11 A0A182UQ73 A0A182U3K2 A0A182X1L6 A0A182IEJ8 A0A182QRK1 A0A0P5ZHM8 A0A240PJZ3 Q2HXW9 A0A182MR29 A0A182XZJ5 A0A2C9H419 A0A1Y9HCG0 A0A1I8JVF3 A0A1I8JV65 A0A182L2F0 A0A1Y9IM13 A0A1I8JT09 Q7Q3D8 A0A182MZE9 T1E772 A0A2M4A0M5 A0A2M4BNJ2 A0A182JPU4 A0A2P0XIF0 A0A1I8JSH0 A0A1Y9IVW2 A0A0P5CXY1 G9C5D8 A0A2L2Y0Q4 A0A182RVL0 A0A1B6GJX3 U5EVU9 A0A151IXY3 A0A0P5T616 T1DC53 A0A0N8CXU9 A0A2M3YZD2 A0A0P5SG68
PDB
5WRO
E-value=2.3748e-130,
Score=1194
Ontologies
PATHWAY
00010
Glycolysis / Gluconeogenesis - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
03018 RNA degradation - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
01230 Biosynthesis of amino acids - Bombyx mori (domestic silkworm)
03018 RNA degradation - Bombyx mori (domestic silkworm)
GO
PANTHER
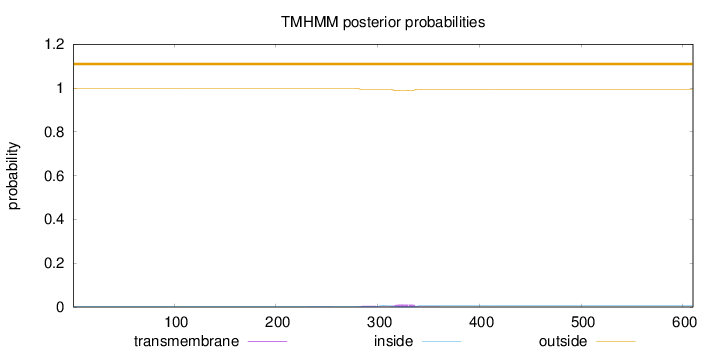
Topology
Length:
610
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.36321
Exp number, first 60 AAs:
0.00184
Total prob of N-in:
0.00237
outside
1 - 610
Population Genetic Test Statistics
Pi
21.600562
Theta
21.352765
Tajima's D
0.164766
CLR
0.037676
CSRT
0.420378981050947
Interpretation
Uncertain
