Gene
KWMTBOMO00492 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000825
Annotation
fatty_alcohol_acetyltransferase_[Agrotis_segetum]
Location in the cell
PlasmaMembrane Reliability : 4.616
Sequence
CDS
ATGTGGGGGATATTTCTATATATTTTGGATCTGGTGGGTCTCAGCCCGATTCCATTTCTGGCAGAATTAATTGGGACTACGGCACCTGCTCTAAAACTACTTATATCGATCCTAATGGGATATCCAATAGCAATACTTTATCATCACCATATCAAAGATTTCTTCGACTACCGAAACATATACTTCATTGTTTGTGGAGTAGATATGGCATTTTTCAATTTTGGAATATCAATGTACCATAATATGATTCCCGCAATTGTTATTTACATGACCACAAAGTTTTTGGGAGCTGGAAAAATGAATGCAGTGATAACATTTGTATTTAATATGGGCTATCTTCTTATTGGTTACATTGTCACAGAGTCTGAAGATTATGATATAACATGGACCATGCCTCACTGTGTTTTGACATTGAAACTGATGGCTCTGTCCTTTGATTTATGGGATGGAAAGAAAATGATCAAAGGCGAGACTCTCTCAGAGAACAATAAGAAGACTGCTTTATTAAGAGCTCCATCTTTTGTAGAATTGATTGGCTTTGTTTACTTTCCTGCCTGCTTCTTAGTTGGACCTATATTTTCGTTCCGGCGATATATTGACTTTGTAATGGACAAGTTTCCTGTTGAGAAAGAAGCAAAGGTGTATGAGAAACAGGCATTAAGGAGATTGCTTCAGGGTTTGGGGTATTTGATTGCTTTTCAAGTGGGAGTTTACGTATTCAATGTGAAATACATGCTATCTGATGATTTCTGGGAGACTTCGATCTTCTATCGGCACTTCTATTGTGGCATGTGGGCACACTTTGCTCTCTATAAGTACATCTCTTGCTGGCTACTCACTGAAGCGTCTTGCATCAGATTCGGGATGTCTTACAACGGCGTCGAGACCACACCGGAAGGTAAGACGATCTCCAAATGGGACGGATGTAACAACATCAAGCTATTGAGATTCGAGGGGGCCACAAAGTTCCAGCACTACATCGACAGCTTCAATTGCAACACGAACCACTTCGCTGCCGAGTACATTTATAAACGACTGAAATTCCTAGGAAACCGCAACCTCAGTCAGTTCTTTACGCTTCTGTTCCTCGCTCTATGGCACGGAACCAGGTCCGGCTATTACATGACCTTCTTCAACGAATTTCTGATAATGCTCATGGAGAAAGAACTGGAACCAATTATTTCTAAAACGGAGCTTTACAACAAACTATGGTCTCACAATGTAACCAAGTACATGCTCTATGGCTTATTGAAAATGTATACAATTGTATTTATGGGCTGGAGTCTTGCTCCATTCGATCTGAAGGCCTTCTCGAAATGGTGGGGAGTTTACTCGAGTTTATATTACTCAGGATTTGTTCTTTTCTTACCGTGGGCCATTTTGTATAAGCCCCTCTTAAAAATGGCTATCAAGGCTTCCGGCGTCCGTCGCGTTAGAGCTGATTAA
Protein
MWGIFLYILDLVGLSPIPFLAELIGTTAPALKLLISILMGYPIAILYHHHIKDFFDYRNIYFIVCGVDMAFFNFGISMYHNMIPAIVIYMTTKFLGAGKMNAVITFVFNMGYLLIGYIVTESEDYDITWTMPHCVLTLKLMALSFDLWDGKKMIKGETLSENNKKTALLRAPSFVELIGFVYFPACFLVGPIFSFRRYIDFVMDKFPVEKEAKVYEKQALRRLLQGLGYLIAFQVGVYVFNVKYMLSDDFWETSIFYRHFYCGMWAHFALYKYISCWLLTEASCIRFGMSYNGVETTPEGKTISKWDGCNNIKLLRFEGATKFQHYIDSFNCNTNHFAAEYIYKRLKFLGNRNLSQFFTLLFLALWHGTRSGYYMTFFNEFLIMLMEKELEPIISKTELYNKLWSHNVTKYMLYGLLKMYTIVFMGWSLAPFDLKAFSKWWGVYSSLYYSGFVLFLPWAILYKPLLKMAIKASGVRRVRAD
Summary
Similarity
Belongs to the membrane-bound acyltransferase family.
Belongs to the globin family.
Belongs to the globin family.
Uniprot
H9IU95
A0A088M9J9
A0A2H1VAA7
A0A1V0M8A4
A0A0P0EM67
A0A291P135
+ More
A0A1L8D692 A0A194QU00 A0A194PEZ6 A0A291P130 A0A0L7KWQ2 A0A212EIA4 S4P468 A0A1B6HRE3 A0A1B6F3T4 A0A1B6FRU6 A0A067QH56 K7IM10 A0A1B6L9D4 A0A1B6KDI7 A0A2J7RDZ6 A0A023EYR8 A0A1W4WF49 A0A232F3H8 A0A0C9QQK1 Q179L9 A0A023EUY0 A0A1Q3FYC6 A0A0V0G6P9 A0A1B6D3K0 A0A1Y1KXF3 A0A1Q3FYK3 A0A1Q3FYH6 A0A1Q3FYR1 A0A1Q3FYE9 A0A069DZ20 A0A1B6KKI3 A0A1B6M2B1 A0A026W0B1 A0A0P4WFQ0 N6U8Z0 T1I5E5 U4U697 E2AU46 A0A0N7Z934 A0A0N7ZAX6 V5G709 A0A182JNX5 A0A2M4BLS1 A0A182Y1V3 A0A182PBP2 A0A182GGP3 A0A182SWW2 W5JMS6 A0A182FTU8 A0A2M3Z7U2 A0A2M3Z7S6 A0A2M4AJ83 A0A182M979 A0A195CVH2 A0A182TKA3 F4WZI6 Q7Q049 A0A2M4AI66 A0A182XD88 A0A182HQ02 A0A151X1T9 A0A087U0R2 A0A0P5A9L6 A0A0P4XT72 A0A0P5ZN09 A0A0N8A2D7 A0A0P5CL10 A0A0P4YBX6 A0A084WI00 A0A0A9ZAQ7 A0A0P5UXX2 A0A0P6B7V6 A0A0P5DLA4 A0A0P5AP74 E9GAM7 A0A2L2Y2V1 A0A0N8DFW9 U5EWS2 A0A0P6I2Q2 A0A182IL59 A0A195ET03 A0A0P4ZDZ5 A0A0P5RXH1 A0A0P6DLW2 A0A2P6KF69 A0A0P5UV56 E9IE62 A0A182QYF4 A0A158NH37 A0A182MYH5 A0A195BVW0 B4H954 A0A182W986
A0A1L8D692 A0A194QU00 A0A194PEZ6 A0A291P130 A0A0L7KWQ2 A0A212EIA4 S4P468 A0A1B6HRE3 A0A1B6F3T4 A0A1B6FRU6 A0A067QH56 K7IM10 A0A1B6L9D4 A0A1B6KDI7 A0A2J7RDZ6 A0A023EYR8 A0A1W4WF49 A0A232F3H8 A0A0C9QQK1 Q179L9 A0A023EUY0 A0A1Q3FYC6 A0A0V0G6P9 A0A1B6D3K0 A0A1Y1KXF3 A0A1Q3FYK3 A0A1Q3FYH6 A0A1Q3FYR1 A0A1Q3FYE9 A0A069DZ20 A0A1B6KKI3 A0A1B6M2B1 A0A026W0B1 A0A0P4WFQ0 N6U8Z0 T1I5E5 U4U697 E2AU46 A0A0N7Z934 A0A0N7ZAX6 V5G709 A0A182JNX5 A0A2M4BLS1 A0A182Y1V3 A0A182PBP2 A0A182GGP3 A0A182SWW2 W5JMS6 A0A182FTU8 A0A2M3Z7U2 A0A2M3Z7S6 A0A2M4AJ83 A0A182M979 A0A195CVH2 A0A182TKA3 F4WZI6 Q7Q049 A0A2M4AI66 A0A182XD88 A0A182HQ02 A0A151X1T9 A0A087U0R2 A0A0P5A9L6 A0A0P4XT72 A0A0P5ZN09 A0A0N8A2D7 A0A0P5CL10 A0A0P4YBX6 A0A084WI00 A0A0A9ZAQ7 A0A0P5UXX2 A0A0P6B7V6 A0A0P5DLA4 A0A0P5AP74 E9GAM7 A0A2L2Y2V1 A0A0N8DFW9 U5EWS2 A0A0P6I2Q2 A0A182IL59 A0A195ET03 A0A0P4ZDZ5 A0A0P5RXH1 A0A0P6DLW2 A0A2P6KF69 A0A0P5UV56 E9IE62 A0A182QYF4 A0A158NH37 A0A182MYH5 A0A195BVW0 B4H954 A0A182W986
Pubmed
19121390
26385554
28349297
26445454
28986331
26354079
+ More
26227816 22118469 23622113 24845553 20075255 25474469 28648823 17510324 24945155 28004739 26334808 24508170 30249741 23537049 20798317 27129103 25244985 26483478 20920257 23761445 21719571 12364791 24438588 25401762 26823975 21292972 26561354 21282665 21347285 17994087
26227816 22118469 23622113 24845553 20075255 25474469 28648823 17510324 24945155 28004739 26334808 24508170 30249741 23537049 20798317 27129103 25244985 26483478 20920257 23761445 21719571 12364791 24438588 25401762 26823975 21292972 26561354 21282665 21347285 17994087
EMBL
BABH01044294
BABH01044295
BABH01044296
KJ579218
AIN34694.1
ODYU01001491
+ More
SOQ37769.1 KU755505 ARD71215.1 KT261729 ALJ30269.1 MF687659 ATJ44585.1 GEYN01000157 JAV01972.1 KQ461108 KPJ09008.1 KQ459605 KPI91852.1 MF706184 ATJ44611.1 JTDY01004989 KOB67501.1 AGBW02014676 OWR41222.1 GAIX01009192 JAA83368.1 GECU01030511 GECU01007131 JAS77195.1 JAT00576.1 GECZ01024847 GECZ01003357 JAS44922.1 JAS66412.1 GECZ01016823 JAS52946.1 KK853408 KDR07796.1 GEBQ01019658 GEBQ01018630 GEBQ01015893 JAT20319.1 JAT21347.1 JAT24084.1 GEBQ01030481 GEBQ01017578 JAT09496.1 JAT22399.1 NEVH01005280 PNF39053.1 GBBI01004938 JAC13774.1 NNAY01001056 OXU25314.1 GBYB01005999 JAG75766.1 CH477348 EAT42940.1 GAPW01001314 JAC12284.1 GFDL01002466 JAV32579.1 GECL01002348 JAP03776.1 GEDC01017031 JAS20267.1 GEZM01076918 JAV63547.1 GFDL01002445 JAV32600.1 GFDL01002410 JAV32635.1 GFDL01002377 JAV32668.1 GFDL01002440 JAV32605.1 GBGD01001405 JAC87484.1 GEBQ01028027 GEBQ01023382 JAT11950.1 JAT16595.1 GEBQ01024098 GEBQ01009939 JAT15879.1 JAT30038.1 KK107519 QOIP01000009 EZA49360.1 RLU18622.1 GDRN01092622 JAI60073.1 APGK01044206 KB741021 ENN75052.1 ACPB03014450 KB632003 ERL87828.1 GL442796 EFN63043.1 GDKW01001659 JAI54936.1 GDRN01092623 JAI60072.1 GALX01002591 JAB65875.1 GGFJ01004750 MBW53891.1 JXUM01062007 JXUM01062008 KQ562178 KXJ76490.1 ADMH02000662 ETN65406.1 GGFM01003845 MBW24596.1 GGFM01003804 MBW24555.1 GGFK01007528 MBW40849.1 AXCM01014929 AXCM01014930 KQ977259 KYN04537.1 GL888479 EGI60430.1 AAAB01008986 EAA00268.4 GGFK01007164 MBW40485.1 APCN01003286 KQ982585 KYQ54381.1 KK117608 KFM70951.1 GDIP01202196 JAJ21206.1 GDIP01237039 JAI86362.1 GDIP01069707 GDIP01054866 LRGB01000781 JAM48849.1 KZS16001.1 GDIP01189241 JAJ34161.1 GDIP01169935 JAJ53467.1 GDIP01229694 JAI93707.1 ATLV01023900 KE525347 KFB49844.1 GBHO01001247 GBRD01008464 GDHC01002671 JAG42357.1 JAG57357.1 JAQ15958.1 GDIP01109998 JAL93716.1 GDIP01018470 JAM85245.1 GDIP01154549 JAJ68853.1 GDIP01199735 JAJ23667.1 GL732537 EFX83278.1 IAAA01009389 LAA02502.1 GDIP01037695 JAM66020.1 GANO01002837 JAB57034.1 GDIQ01010465 JAN84272.1 KQ981979 KYN31380.1 GDIP01215595 JAJ07807.1 GDIQ01094989 JAL56737.1 GDIQ01090399 JAN04338.1 MWRG01013705 PRD24951.1 GDIP01107898 JAL95816.1 GL762556 EFZ21225.1 AXCN02001044 ADTU01001624 ADTU01001625 KQ976398 KYM92769.1 CH479227 EDW35278.1
SOQ37769.1 KU755505 ARD71215.1 KT261729 ALJ30269.1 MF687659 ATJ44585.1 GEYN01000157 JAV01972.1 KQ461108 KPJ09008.1 KQ459605 KPI91852.1 MF706184 ATJ44611.1 JTDY01004989 KOB67501.1 AGBW02014676 OWR41222.1 GAIX01009192 JAA83368.1 GECU01030511 GECU01007131 JAS77195.1 JAT00576.1 GECZ01024847 GECZ01003357 JAS44922.1 JAS66412.1 GECZ01016823 JAS52946.1 KK853408 KDR07796.1 GEBQ01019658 GEBQ01018630 GEBQ01015893 JAT20319.1 JAT21347.1 JAT24084.1 GEBQ01030481 GEBQ01017578 JAT09496.1 JAT22399.1 NEVH01005280 PNF39053.1 GBBI01004938 JAC13774.1 NNAY01001056 OXU25314.1 GBYB01005999 JAG75766.1 CH477348 EAT42940.1 GAPW01001314 JAC12284.1 GFDL01002466 JAV32579.1 GECL01002348 JAP03776.1 GEDC01017031 JAS20267.1 GEZM01076918 JAV63547.1 GFDL01002445 JAV32600.1 GFDL01002410 JAV32635.1 GFDL01002377 JAV32668.1 GFDL01002440 JAV32605.1 GBGD01001405 JAC87484.1 GEBQ01028027 GEBQ01023382 JAT11950.1 JAT16595.1 GEBQ01024098 GEBQ01009939 JAT15879.1 JAT30038.1 KK107519 QOIP01000009 EZA49360.1 RLU18622.1 GDRN01092622 JAI60073.1 APGK01044206 KB741021 ENN75052.1 ACPB03014450 KB632003 ERL87828.1 GL442796 EFN63043.1 GDKW01001659 JAI54936.1 GDRN01092623 JAI60072.1 GALX01002591 JAB65875.1 GGFJ01004750 MBW53891.1 JXUM01062007 JXUM01062008 KQ562178 KXJ76490.1 ADMH02000662 ETN65406.1 GGFM01003845 MBW24596.1 GGFM01003804 MBW24555.1 GGFK01007528 MBW40849.1 AXCM01014929 AXCM01014930 KQ977259 KYN04537.1 GL888479 EGI60430.1 AAAB01008986 EAA00268.4 GGFK01007164 MBW40485.1 APCN01003286 KQ982585 KYQ54381.1 KK117608 KFM70951.1 GDIP01202196 JAJ21206.1 GDIP01237039 JAI86362.1 GDIP01069707 GDIP01054866 LRGB01000781 JAM48849.1 KZS16001.1 GDIP01189241 JAJ34161.1 GDIP01169935 JAJ53467.1 GDIP01229694 JAI93707.1 ATLV01023900 KE525347 KFB49844.1 GBHO01001247 GBRD01008464 GDHC01002671 JAG42357.1 JAG57357.1 JAQ15958.1 GDIP01109998 JAL93716.1 GDIP01018470 JAM85245.1 GDIP01154549 JAJ68853.1 GDIP01199735 JAJ23667.1 GL732537 EFX83278.1 IAAA01009389 LAA02502.1 GDIP01037695 JAM66020.1 GANO01002837 JAB57034.1 GDIQ01010465 JAN84272.1 KQ981979 KYN31380.1 GDIP01215595 JAJ07807.1 GDIQ01094989 JAL56737.1 GDIQ01090399 JAN04338.1 MWRG01013705 PRD24951.1 GDIP01107898 JAL95816.1 GL762556 EFZ21225.1 AXCN02001044 ADTU01001624 ADTU01001625 KQ976398 KYM92769.1 CH479227 EDW35278.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000037510
UP000007151
UP000027135
+ More
UP000002358 UP000235965 UP000192223 UP000215335 UP000008820 UP000053097 UP000279307 UP000019118 UP000015103 UP000030742 UP000000311 UP000075881 UP000076408 UP000075885 UP000069940 UP000249989 UP000075901 UP000000673 UP000069272 UP000075883 UP000078542 UP000075902 UP000007755 UP000007062 UP000076407 UP000075840 UP000075809 UP000054359 UP000076858 UP000030765 UP000000305 UP000075880 UP000078541 UP000075886 UP000005205 UP000075884 UP000078540 UP000008744 UP000075920
UP000002358 UP000235965 UP000192223 UP000215335 UP000008820 UP000053097 UP000279307 UP000019118 UP000015103 UP000030742 UP000000311 UP000075881 UP000076408 UP000075885 UP000069940 UP000249989 UP000075901 UP000000673 UP000069272 UP000075883 UP000078542 UP000075902 UP000007755 UP000007062 UP000076407 UP000075840 UP000075809 UP000054359 UP000076858 UP000030765 UP000000305 UP000075880 UP000078541 UP000075886 UP000005205 UP000075884 UP000078540 UP000008744 UP000075920
PRIDE
Interpro
SUPFAM
SSF46458
SSF46458
Gene 3D
ProteinModelPortal
H9IU95
A0A088M9J9
A0A2H1VAA7
A0A1V0M8A4
A0A0P0EM67
A0A291P135
+ More
A0A1L8D692 A0A194QU00 A0A194PEZ6 A0A291P130 A0A0L7KWQ2 A0A212EIA4 S4P468 A0A1B6HRE3 A0A1B6F3T4 A0A1B6FRU6 A0A067QH56 K7IM10 A0A1B6L9D4 A0A1B6KDI7 A0A2J7RDZ6 A0A023EYR8 A0A1W4WF49 A0A232F3H8 A0A0C9QQK1 Q179L9 A0A023EUY0 A0A1Q3FYC6 A0A0V0G6P9 A0A1B6D3K0 A0A1Y1KXF3 A0A1Q3FYK3 A0A1Q3FYH6 A0A1Q3FYR1 A0A1Q3FYE9 A0A069DZ20 A0A1B6KKI3 A0A1B6M2B1 A0A026W0B1 A0A0P4WFQ0 N6U8Z0 T1I5E5 U4U697 E2AU46 A0A0N7Z934 A0A0N7ZAX6 V5G709 A0A182JNX5 A0A2M4BLS1 A0A182Y1V3 A0A182PBP2 A0A182GGP3 A0A182SWW2 W5JMS6 A0A182FTU8 A0A2M3Z7U2 A0A2M3Z7S6 A0A2M4AJ83 A0A182M979 A0A195CVH2 A0A182TKA3 F4WZI6 Q7Q049 A0A2M4AI66 A0A182XD88 A0A182HQ02 A0A151X1T9 A0A087U0R2 A0A0P5A9L6 A0A0P4XT72 A0A0P5ZN09 A0A0N8A2D7 A0A0P5CL10 A0A0P4YBX6 A0A084WI00 A0A0A9ZAQ7 A0A0P5UXX2 A0A0P6B7V6 A0A0P5DLA4 A0A0P5AP74 E9GAM7 A0A2L2Y2V1 A0A0N8DFW9 U5EWS2 A0A0P6I2Q2 A0A182IL59 A0A195ET03 A0A0P4ZDZ5 A0A0P5RXH1 A0A0P6DLW2 A0A2P6KF69 A0A0P5UV56 E9IE62 A0A182QYF4 A0A158NH37 A0A182MYH5 A0A195BVW0 B4H954 A0A182W986
A0A1L8D692 A0A194QU00 A0A194PEZ6 A0A291P130 A0A0L7KWQ2 A0A212EIA4 S4P468 A0A1B6HRE3 A0A1B6F3T4 A0A1B6FRU6 A0A067QH56 K7IM10 A0A1B6L9D4 A0A1B6KDI7 A0A2J7RDZ6 A0A023EYR8 A0A1W4WF49 A0A232F3H8 A0A0C9QQK1 Q179L9 A0A023EUY0 A0A1Q3FYC6 A0A0V0G6P9 A0A1B6D3K0 A0A1Y1KXF3 A0A1Q3FYK3 A0A1Q3FYH6 A0A1Q3FYR1 A0A1Q3FYE9 A0A069DZ20 A0A1B6KKI3 A0A1B6M2B1 A0A026W0B1 A0A0P4WFQ0 N6U8Z0 T1I5E5 U4U697 E2AU46 A0A0N7Z934 A0A0N7ZAX6 V5G709 A0A182JNX5 A0A2M4BLS1 A0A182Y1V3 A0A182PBP2 A0A182GGP3 A0A182SWW2 W5JMS6 A0A182FTU8 A0A2M3Z7U2 A0A2M3Z7S6 A0A2M4AJ83 A0A182M979 A0A195CVH2 A0A182TKA3 F4WZI6 Q7Q049 A0A2M4AI66 A0A182XD88 A0A182HQ02 A0A151X1T9 A0A087U0R2 A0A0P5A9L6 A0A0P4XT72 A0A0P5ZN09 A0A0N8A2D7 A0A0P5CL10 A0A0P4YBX6 A0A084WI00 A0A0A9ZAQ7 A0A0P5UXX2 A0A0P6B7V6 A0A0P5DLA4 A0A0P5AP74 E9GAM7 A0A2L2Y2V1 A0A0N8DFW9 U5EWS2 A0A0P6I2Q2 A0A182IL59 A0A195ET03 A0A0P4ZDZ5 A0A0P5RXH1 A0A0P6DLW2 A0A2P6KF69 A0A0P5UV56 E9IE62 A0A182QYF4 A0A158NH37 A0A182MYH5 A0A195BVW0 B4H954 A0A182W986
Ontologies
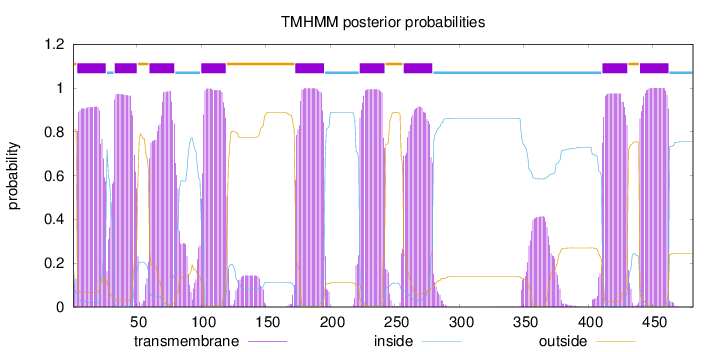
Topology
Length:
481
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
198.11193
Exp number, first 60 AAs:
39.93561
Total prob of N-in:
0.19097
POSSIBLE N-term signal
sequence
outside
1 - 3
TMhelix
4 - 26
inside
27 - 32
TMhelix
33 - 50
outside
51 - 59
TMhelix
60 - 79
inside
80 - 99
TMhelix
100 - 119
outside
120 - 172
TMhelix
173 - 195
inside
196 - 222
TMhelix
223 - 242
outside
243 - 256
TMhelix
257 - 279
inside
280 - 410
TMhelix
411 - 430
outside
431 - 439
TMhelix
440 - 462
inside
463 - 481
Population Genetic Test Statistics
Pi
25.367477
Theta
25.344153
Tajima's D
-0.604688
CLR
0.014706
CSRT
0.218639068046598
Interpretation
Uncertain
