Pre Gene Modal
BGIBMGA012292
Annotation
PREDICTED:_ubiquitin_carboxyl-terminal_hydrolase_isozyme_L5_isoform_X2_[Bombyx_mori]
Full name
Ubiquitin carboxyl-terminal hydrolase
Location in the cell
Cytoplasmic Reliability : 3.021
Sequence
CDS
ATGGATTCTGCCGGGGATTGGTGTCTCCTAGAGAGTGATCCTGGAGTTTTCACTCAGCTAATTCACAAATTTGGTGCCAAAGGTGTACAAGTTGAGGAACTGTGGACTATTGAAGACAGCATGTTTGAAAATTTGAGACCAGTGCATGGTCTCATTTTCCTGTTCAAGTATGTTAATTATGAAGAACCCGCTGGGCCTATTGTATCAGATGATCGCTTAGACAAAATATTTTTTGCCAAACAGGTTATCAATAATGCCTGCGCAACTCAAGCTGTAATCAGTCTCCTGTTAAATTGCAAACACCCAGATTTGGAACTTGGTCCTGAATTGACCAAACTGAAAGAATTCAGCATGACATTCGACTCGCGCATGAGAGGTCTCTCGTTAAGCAACTCACAGACTATACGCAGCGCACACAACTCAATGGCTCAACAGCCTCTCTTTGAAGTAGATCCTAAAATGCCTGCCAAAGATGAAGACGCCTATCATTTCATAGGATATATGCCAATTGATGGCCGTCTTTACGAACTGGATGGGCTACAAGAAGGTCCGATTGATCATGGTGCAGTTGCTCCGGAACAAGATTGGCTCGATGTTGTCCGTCCAATTATAATGAGAAGGATTAATGTATACACCGAAGGAGAAATTCACTTCAACTTGATGGCCCTCGTATCAGATCGTAAAATGATATATCAACGTCAGCTTGATACACTTCTTAATGAAACACAGTTGCTGGGTATGGAAACCGATGCTCTCGATACAGAAATAGCGAGACTGAGAATGTTGATTGATTATGAGGAAATAAAGATGGCAAAGTACAAACAGGAGTTGGTACGCAGGCGTCACAATTACATGCCGTTCATCATTAATTTGCTTCAAATACTGGCTAAGGAGAAGAAGCTGATGCCCCTGCTGGAGAAGGCCAAGGAGCGCGCTACCAAGCGGAGTCACAAGAAAATGAAAGTATAA
Protein
MDSAGDWCLLESDPGVFTQLIHKFGAKGVQVEELWTIEDSMFENLRPVHGLIFLFKYVNYEEPAGPIVSDDRLDKIFFAKQVINNACATQAVISLLLNCKHPDLELGPELTKLKEFSMTFDSRMRGLSLSNSQTIRSAHNSMAQQPLFEVDPKMPAKDEDAYHFIGYMPIDGRLYELDGLQEGPIDHGAVAPEQDWLDVVRPIIMRRINVYTEGEIHFNLMALVSDRKMIYQRQLDTLLNETQLLGMETDALDTEIARLRMLIDYEEIKMAKYKQELVRRRHNYMPFIINLLQILAKEKKLMPLLEKAKERATKRSHKKMKV
Summary
Catalytic Activity
Thiol-dependent hydrolysis of ester, thioester, amide, peptide and isopeptide bonds formed by the C-terminal Gly of ubiquitin (a 76-residue protein attached to proteins as an intracellular targeting signal).
Similarity
Belongs to the peptidase C12 family.
Uniprot
H9JRY2
A0A2H1VAA4
A0A212EQG4
A0A1E1WV99
A0A194QVL2
A0A2A4IUH2
+ More
B2DBN6 A0A194PL71 A0A2J7QYF7 A0A182GIG1 U5EXX5 A0A1B6LFK0 A0A1B6CKN2 A0A023EQX2 D6WHM3 Q16RF6 A0A182PP48 A0A2R7WCP0 E0W0S3 A0A2P8Z3T0 A0A084WKJ6 B0WAM9 A0A293MUB2 W5JCU7 A0A2M3ZB04 A0A182SLF3 A0A0L0C8L0 A0A2M4AFG9 A0A1Q3F679 A0A182Y109 A0A182TNW3 A0A182LJF6 Q7Q728 A0A1Y1JTL2 A0A182HLZ2 V3ZUI0 A0A131XCB0 A0A2M4BU01 A0A224Z7H3 A0A182VCC7 A0A131ZA60 A0A182M0Q8 A0A1I8NPS9 A0A0V0G4Z8 T1DM54 A0A182WQW7 L7M3C8 A0A182X543 A0A182JY24 A0A182NM32 A0A067R6D1 A0A0P4X1K4 A0A0T6B272 A0A182R0J9 T1PNQ0 A0A1I8MJB6 B9VRB6 R4FLL1 A0A0K8TNJ7 A0A0K8U9T8 A0A069DSS4 A0A034WE06 A0A023FA18 A0A1A9WTA5 A0A224Z7E3 A0A0A1WPP9 A0A224XTE9 A0A023GKP6 G3MJU9 E9FUJ0 A0A131XTV2 B7P6T3 A0A2R5L8K1 K7J0N8 W8BQH6 A0A232F3R8 A0A3B4A0M5 A0A385MQ64 A0A0K8RE55 A0A1S3I7D4 A0A023FLF1 C1BIU4 A0A3P8ZFF2 A0A1A9Y047 A0A1A9ZKG1 H3D466 A0A1A7WMN5 A0A1A9UG24 A0A1B0BC56 A0A1A8KNB6 A0A1A8SE78 A0A1A8Q2R0 A0A1A8CXK7 A0A1A8AX75 A0A3B5KHX8 D3TPP8 A0A3B4ES32 A0A3P8R871 A0A3P9CGK5 A0A146P5D3
B2DBN6 A0A194PL71 A0A2J7QYF7 A0A182GIG1 U5EXX5 A0A1B6LFK0 A0A1B6CKN2 A0A023EQX2 D6WHM3 Q16RF6 A0A182PP48 A0A2R7WCP0 E0W0S3 A0A2P8Z3T0 A0A084WKJ6 B0WAM9 A0A293MUB2 W5JCU7 A0A2M3ZB04 A0A182SLF3 A0A0L0C8L0 A0A2M4AFG9 A0A1Q3F679 A0A182Y109 A0A182TNW3 A0A182LJF6 Q7Q728 A0A1Y1JTL2 A0A182HLZ2 V3ZUI0 A0A131XCB0 A0A2M4BU01 A0A224Z7H3 A0A182VCC7 A0A131ZA60 A0A182M0Q8 A0A1I8NPS9 A0A0V0G4Z8 T1DM54 A0A182WQW7 L7M3C8 A0A182X543 A0A182JY24 A0A182NM32 A0A067R6D1 A0A0P4X1K4 A0A0T6B272 A0A182R0J9 T1PNQ0 A0A1I8MJB6 B9VRB6 R4FLL1 A0A0K8TNJ7 A0A0K8U9T8 A0A069DSS4 A0A034WE06 A0A023FA18 A0A1A9WTA5 A0A224Z7E3 A0A0A1WPP9 A0A224XTE9 A0A023GKP6 G3MJU9 E9FUJ0 A0A131XTV2 B7P6T3 A0A2R5L8K1 K7J0N8 W8BQH6 A0A232F3R8 A0A3B4A0M5 A0A385MQ64 A0A0K8RE55 A0A1S3I7D4 A0A023FLF1 C1BIU4 A0A3P8ZFF2 A0A1A9Y047 A0A1A9ZKG1 H3D466 A0A1A7WMN5 A0A1A9UG24 A0A1B0BC56 A0A1A8KNB6 A0A1A8SE78 A0A1A8Q2R0 A0A1A8CXK7 A0A1A8AX75 A0A3B5KHX8 D3TPP8 A0A3B4ES32 A0A3P8R871 A0A3P9CGK5 A0A146P5D3
EC Number
3.4.19.12
Pubmed
19121390
22118469
26354079
18712529
26483478
24945155
+ More
18362917 19820115 17510324 20566863 29403074 24438588 20920257 23761445 26108605 25244985 20966253 12364791 14747013 17210077 28004739 23254933 28049606 28797301 26830274 25576852 24845553 25315136 26369729 26334808 25348373 25474469 25830018 22216098 21292972 20075255 24495485 28648823 25463417 25069045 15496914 21551351 20353571 25186727
18362917 19820115 17510324 20566863 29403074 24438588 20920257 23761445 26108605 25244985 20966253 12364791 14747013 17210077 28004739 23254933 28049606 28797301 26830274 25576852 24845553 25315136 26369729 26334808 25348373 25474469 25830018 22216098 21292972 20075255 24495485 28648823 25463417 25069045 15496914 21551351 20353571 25186727
EMBL
BABH01016939
BABH01016940
ODYU01001491
SOQ37768.1
AGBW02013269
OWR43726.1
+ More
GDQN01004867 GDQN01000114 JAT86187.1 JAT90940.1 KQ461108 KPJ09005.1 NWSH01006566 PCG63361.1 AB264721 BAG30795.1 KQ459605 KPI91855.1 NEVH01009093 PNF33612.1 JXUM01011979 JXUM01030441 KQ560885 KQ560342 KXJ80478.1 KXJ82952.1 GANO01000720 JAB59151.1 GEBQ01017511 JAT22466.1 GEDC01023259 JAS14039.1 GAPW01002577 JAC11021.1 KQ971331 EEZ99725.1 CH477712 EAT36993.1 KK854627 PTY17443.1 AAZO01006830 DS235862 EEB19229.1 PYGN01000208 PSN51161.1 ATLV01024123 KE525349 KFB50740.1 DS231873 EDS41583.1 GFWV01019674 MAA44402.1 ADMH02001883 ETN60654.1 GGFM01004958 MBW25709.1 JRES01000850 KNC27734.1 GGFK01006205 MBW39526.1 GFDL01011975 JAV23070.1 AAAB01008960 EAA11802.2 GEZM01101438 JAV52654.1 APCN01004280 KB203470 ESO84586.1 GEFH01004569 JAP64012.1 GGFJ01007388 MBW56529.1 GFPF01013083 MAA24229.1 GEDV01000852 JAP87705.1 AXCM01005810 GECL01002985 JAP03139.1 GAMD01000081 JAB01510.1 GACK01007401 JAA57633.1 KK852714 KDR17906.1 GDRN01027813 JAI67634.1 LJIG01016121 KRT81551.1 AXCN02002327 KA647066 KA649713 KA649715 AFP64344.1 FJ595021 ACM43511.1 ACPB03013647 GAHY01001338 JAA76172.1 GDAI01001664 JAI15939.1 GDHF01028966 JAI23348.1 GBGD01002167 JAC86722.1 GAKP01006028 JAC52924.1 GBBI01000868 JAC17844.1 GFPF01013043 MAA24189.1 GBXI01013797 JAD00495.1 GFTR01005043 JAW11383.1 GBBM01001720 JAC33698.1 JO842150 AEO33767.1 GL732525 EFX88738.1 GEFM01006795 JAP69001.1 ABJB011067020 ABJB011080114 DS647752 EEC02305.1 GGLE01001698 MBY05824.1 GAMC01003075 JAC03481.1 NNAY01001035 OXU25401.1 MG462699 AYA62525.1 GADI01004378 JAA69430.1 GBBK01002989 JAC21493.1 BT074523 ACO08947.1 HADW01005822 HADX01009792 SBP07222.1 JXJN01011850 HAEE01013693 SBR33743.1 HAEH01014265 HAEI01013490 SBS15959.1 HAEF01011676 HAEG01010689 SBR87786.1 HADZ01019624 HAEA01001632 SBP83565.1 HADY01020586 HAEJ01004709 SBP59071.1 CCAG010023540 EZ423400 ADD19676.1 GCES01147285 GCES01146836 GCES01003729 JAQ39037.1
GDQN01004867 GDQN01000114 JAT86187.1 JAT90940.1 KQ461108 KPJ09005.1 NWSH01006566 PCG63361.1 AB264721 BAG30795.1 KQ459605 KPI91855.1 NEVH01009093 PNF33612.1 JXUM01011979 JXUM01030441 KQ560885 KQ560342 KXJ80478.1 KXJ82952.1 GANO01000720 JAB59151.1 GEBQ01017511 JAT22466.1 GEDC01023259 JAS14039.1 GAPW01002577 JAC11021.1 KQ971331 EEZ99725.1 CH477712 EAT36993.1 KK854627 PTY17443.1 AAZO01006830 DS235862 EEB19229.1 PYGN01000208 PSN51161.1 ATLV01024123 KE525349 KFB50740.1 DS231873 EDS41583.1 GFWV01019674 MAA44402.1 ADMH02001883 ETN60654.1 GGFM01004958 MBW25709.1 JRES01000850 KNC27734.1 GGFK01006205 MBW39526.1 GFDL01011975 JAV23070.1 AAAB01008960 EAA11802.2 GEZM01101438 JAV52654.1 APCN01004280 KB203470 ESO84586.1 GEFH01004569 JAP64012.1 GGFJ01007388 MBW56529.1 GFPF01013083 MAA24229.1 GEDV01000852 JAP87705.1 AXCM01005810 GECL01002985 JAP03139.1 GAMD01000081 JAB01510.1 GACK01007401 JAA57633.1 KK852714 KDR17906.1 GDRN01027813 JAI67634.1 LJIG01016121 KRT81551.1 AXCN02002327 KA647066 KA649713 KA649715 AFP64344.1 FJ595021 ACM43511.1 ACPB03013647 GAHY01001338 JAA76172.1 GDAI01001664 JAI15939.1 GDHF01028966 JAI23348.1 GBGD01002167 JAC86722.1 GAKP01006028 JAC52924.1 GBBI01000868 JAC17844.1 GFPF01013043 MAA24189.1 GBXI01013797 JAD00495.1 GFTR01005043 JAW11383.1 GBBM01001720 JAC33698.1 JO842150 AEO33767.1 GL732525 EFX88738.1 GEFM01006795 JAP69001.1 ABJB011067020 ABJB011080114 DS647752 EEC02305.1 GGLE01001698 MBY05824.1 GAMC01003075 JAC03481.1 NNAY01001035 OXU25401.1 MG462699 AYA62525.1 GADI01004378 JAA69430.1 GBBK01002989 JAC21493.1 BT074523 ACO08947.1 HADW01005822 HADX01009792 SBP07222.1 JXJN01011850 HAEE01013693 SBR33743.1 HAEH01014265 HAEI01013490 SBS15959.1 HAEF01011676 HAEG01010689 SBR87786.1 HADZ01019624 HAEA01001632 SBP83565.1 HADY01020586 HAEJ01004709 SBP59071.1 CCAG010023540 EZ423400 ADD19676.1 GCES01147285 GCES01146836 GCES01003729 JAQ39037.1
Proteomes
UP000005204
UP000007151
UP000053240
UP000218220
UP000053268
UP000235965
+ More
UP000069940 UP000249989 UP000007266 UP000008820 UP000075885 UP000009046 UP000245037 UP000030765 UP000002320 UP000000673 UP000075901 UP000037069 UP000076408 UP000075902 UP000075882 UP000007062 UP000075840 UP000030746 UP000075903 UP000075883 UP000095300 UP000075920 UP000076407 UP000075881 UP000075884 UP000027135 UP000075886 UP000095301 UP000015103 UP000091820 UP000000305 UP000001555 UP000002358 UP000215335 UP000261520 UP000085678 UP000265140 UP000092443 UP000092445 UP000007303 UP000078200 UP000092460 UP000005226 UP000092444 UP000261460 UP000265100 UP000265160
UP000069940 UP000249989 UP000007266 UP000008820 UP000075885 UP000009046 UP000245037 UP000030765 UP000002320 UP000000673 UP000075901 UP000037069 UP000076408 UP000075902 UP000075882 UP000007062 UP000075840 UP000030746 UP000075903 UP000075883 UP000095300 UP000075920 UP000076407 UP000075881 UP000075884 UP000027135 UP000075886 UP000095301 UP000015103 UP000091820 UP000000305 UP000001555 UP000002358 UP000215335 UP000261520 UP000085678 UP000265140 UP000092443 UP000092445 UP000007303 UP000078200 UP000092460 UP000005226 UP000092444 UP000261460 UP000265100 UP000265160
PRIDE
Interpro
SUPFAM
SSF54001
SSF54001
Gene 3D
ProteinModelPortal
H9JRY2
A0A2H1VAA4
A0A212EQG4
A0A1E1WV99
A0A194QVL2
A0A2A4IUH2
+ More
B2DBN6 A0A194PL71 A0A2J7QYF7 A0A182GIG1 U5EXX5 A0A1B6LFK0 A0A1B6CKN2 A0A023EQX2 D6WHM3 Q16RF6 A0A182PP48 A0A2R7WCP0 E0W0S3 A0A2P8Z3T0 A0A084WKJ6 B0WAM9 A0A293MUB2 W5JCU7 A0A2M3ZB04 A0A182SLF3 A0A0L0C8L0 A0A2M4AFG9 A0A1Q3F679 A0A182Y109 A0A182TNW3 A0A182LJF6 Q7Q728 A0A1Y1JTL2 A0A182HLZ2 V3ZUI0 A0A131XCB0 A0A2M4BU01 A0A224Z7H3 A0A182VCC7 A0A131ZA60 A0A182M0Q8 A0A1I8NPS9 A0A0V0G4Z8 T1DM54 A0A182WQW7 L7M3C8 A0A182X543 A0A182JY24 A0A182NM32 A0A067R6D1 A0A0P4X1K4 A0A0T6B272 A0A182R0J9 T1PNQ0 A0A1I8MJB6 B9VRB6 R4FLL1 A0A0K8TNJ7 A0A0K8U9T8 A0A069DSS4 A0A034WE06 A0A023FA18 A0A1A9WTA5 A0A224Z7E3 A0A0A1WPP9 A0A224XTE9 A0A023GKP6 G3MJU9 E9FUJ0 A0A131XTV2 B7P6T3 A0A2R5L8K1 K7J0N8 W8BQH6 A0A232F3R8 A0A3B4A0M5 A0A385MQ64 A0A0K8RE55 A0A1S3I7D4 A0A023FLF1 C1BIU4 A0A3P8ZFF2 A0A1A9Y047 A0A1A9ZKG1 H3D466 A0A1A7WMN5 A0A1A9UG24 A0A1B0BC56 A0A1A8KNB6 A0A1A8SE78 A0A1A8Q2R0 A0A1A8CXK7 A0A1A8AX75 A0A3B5KHX8 D3TPP8 A0A3B4ES32 A0A3P8R871 A0A3P9CGK5 A0A146P5D3
B2DBN6 A0A194PL71 A0A2J7QYF7 A0A182GIG1 U5EXX5 A0A1B6LFK0 A0A1B6CKN2 A0A023EQX2 D6WHM3 Q16RF6 A0A182PP48 A0A2R7WCP0 E0W0S3 A0A2P8Z3T0 A0A084WKJ6 B0WAM9 A0A293MUB2 W5JCU7 A0A2M3ZB04 A0A182SLF3 A0A0L0C8L0 A0A2M4AFG9 A0A1Q3F679 A0A182Y109 A0A182TNW3 A0A182LJF6 Q7Q728 A0A1Y1JTL2 A0A182HLZ2 V3ZUI0 A0A131XCB0 A0A2M4BU01 A0A224Z7H3 A0A182VCC7 A0A131ZA60 A0A182M0Q8 A0A1I8NPS9 A0A0V0G4Z8 T1DM54 A0A182WQW7 L7M3C8 A0A182X543 A0A182JY24 A0A182NM32 A0A067R6D1 A0A0P4X1K4 A0A0T6B272 A0A182R0J9 T1PNQ0 A0A1I8MJB6 B9VRB6 R4FLL1 A0A0K8TNJ7 A0A0K8U9T8 A0A069DSS4 A0A034WE06 A0A023FA18 A0A1A9WTA5 A0A224Z7E3 A0A0A1WPP9 A0A224XTE9 A0A023GKP6 G3MJU9 E9FUJ0 A0A131XTV2 B7P6T3 A0A2R5L8K1 K7J0N8 W8BQH6 A0A232F3R8 A0A3B4A0M5 A0A385MQ64 A0A0K8RE55 A0A1S3I7D4 A0A023FLF1 C1BIU4 A0A3P8ZFF2 A0A1A9Y047 A0A1A9ZKG1 H3D466 A0A1A7WMN5 A0A1A9UG24 A0A1B0BC56 A0A1A8KNB6 A0A1A8SE78 A0A1A8Q2R0 A0A1A8CXK7 A0A1A8AX75 A0A3B5KHX8 D3TPP8 A0A3B4ES32 A0A3P8R871 A0A3P9CGK5 A0A146P5D3
PDB
4WLR
E-value=1.28274e-107,
Score=995
Ontologies
GO
PANTHER
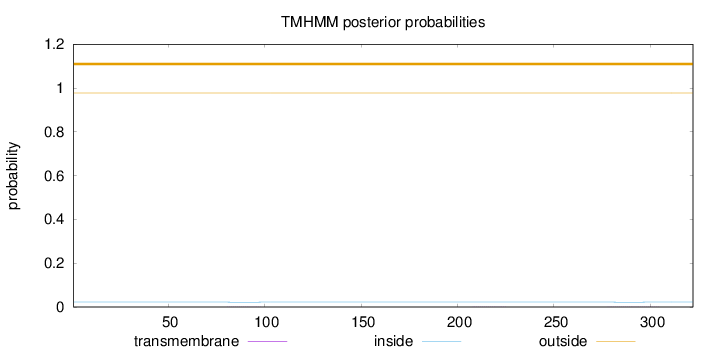
Topology
Length:
322
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.01791
Exp number, first 60 AAs:
0.00041
Total prob of N-in:
0.02234
outside
1 - 322
Population Genetic Test Statistics
Pi
22.864515
Theta
22.457961
Tajima's D
-0.917705
CLR
0.053493
CSRT
0.155242237888106
Interpretation
Uncertain
