Gene
KWMTBOMO00486 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA012295
Annotation
Cdc42_[Spodoptera_litura]
Full name
Cdc42 homolog
Alternative Name
25 kDa GTP-binding protein
Location in the cell
Cytoplasmic Reliability : 1.663 Extracellular Reliability : 1.988
Sequence
CDS
ATGCAGACAATAAAATGTGTTGTTGTGGGTGATGGTGCTGTTGGTAAAACATGTCTTCTGATCAGCTATACAACTAACAAATTTCCATCAGAGTATGTTCCAACTGTATTTGATAACTACGCAGTCACTGTCATGATTGGAGGTGAACCTTACACTCTAGGTCTTTTTGATACTGCAGGTCAGGAAGATTATGACAGGCTAAGACCACTGAGTTATCCTCAAACAGATGTTTTCCTTGTATGCTTCAGTGTTGTGAGCCCTAGTTCATTTGAAAATGTAAAGGAAAAGTGGGTCCCTGAAATAACTCATCATCAACAGAAGACACCGTTTTTGTTGGTTGGGACGCAAATAGACTTGCGCGATGATTCGGCTACAATGGAGAAACTTGCTAAAATAAAACAGAAGCCGGTTTCATTGGAACAAGGAGAGAAATTGGCAAAAGAGCTTAAGGCTGTCAAATATGTAGAATGCTCAGCGCTTACCCAGAAAGGTTTAAAGAACGTGTTCGACGAGGCTATACTTGCTGCACTGGAGCCTCCGGAACCAGCAAAGAAAAAGAAGTGTGTCCTTTTGTAA
Protein
MQTIKCVVVGDGAVGKTCLLISYTTNKFPSEYVPTVFDNYAVTVMIGGEPYTLGLFDTAGQEDYDRLRPLSYPQTDVFLVCFSVVSPSSFENVKEKWVPEITHHQQKTPFLLVGTQIDLRDDSATMEKLAKIKQKPVSLEQGEKLAKELKAVKYVECSALTQKGLKNVFDEAILAALEPPEPAKKKKCVLL
Summary
Description
Regulates mbt kinase activity and is also required to recruit mbt to adherens junctions. Together with mbt, regulates photoreceptor cell morphogenesis (By similarity).
Regulates mbt kinase activity and is also required to recruit mbt to adherens junctions (PubMed:12490550). Together with mbt and Frl, regulates photoreceptor cell morphogenesis (PubMed:12490550, PubMed:26801180). Together with Frl, has a role in the neuronal development of mushroom bodies (PubMed:26801180).
Regulates mbt kinase activity and is also required to recruit mbt to adherens junctions (PubMed:12490550). Together with mbt and Frl, regulates photoreceptor cell morphogenesis (PubMed:12490550, PubMed:26801180). Together with Frl, has a role in the neuronal development of mushroom bodies (PubMed:26801180).
Subunit
Interacts with Frl (via GBD/FH3 domain); the interaction is stronger with the GTP bound form of Cdc42 (PubMed:26801180). The GTP-bound but not the GDP-bound form interacts with mbt and gek (PubMed:9371783, PubMed:12490550).
Similarity
Belongs to the small GTPase superfamily. Rho family. CDC42 subfamily.
Keywords
Cell junction
Cell membrane
Complete proteome
Developmental protein
GTP-binding
Lipoprotein
Membrane
Methylation
Nucleotide-binding
Prenylation
Reference proteome
Feature
chain Cdc42 homolog
propeptide Removed in mature form
propeptide Removed in mature form
Uniprot
H9JRY5
A0A2A4J3X5
A0A2H1WKE4
A0A059VBH0
A0A1E1W6W5
A0A194QU25
+ More
A0A194PL43 A0A212EQJ9 E0VSX8 A0A0L7RAR9 R4WRF3 A0A067R1U4 A0A310SD05 A0A165VG29 A0A2A3E1I2 V5GKL4 N6TXA1 D6WN12 A0A2J7PTR6 A0A1B6GPR3 A0A1B6KK31 A0A1B6J2Q1 A0A1B6FTP5 A0A0K8VS38 A0A023GFP5 G3MKJ3 A0A1E1XJY3 A0A023FF98 L7M3T4 A0A023FUE4 A0A1E1WZ55 A0A224YT61 A0A131Z5G9 A0A131XMD5 W8AM21 A0A034WPG8 A0A0A1WMH4 A0A1W4W8W6 B7PHY7 A0A1B6LTH8 A0A154PP51 A0A0B4J2U1 A0A1S3CZK2 A0A195BRR0 A0A182XWZ5 A0A2M3ZGJ9 A0A2M4A9C9 A0A1Y1KT91 A0A182MGD6 A0A182QIQ5 A0A182VG73 W5JCL2 A0A182NFH5 A0A182K8T7 A0A182UJ57 A0A084VU61 A0A182J293 A0A182LF64 A0A182XI45 F5HK45 A0A182R9Y3 A0A182HT27 Q17031 A0A1A9YJ86 A0A1A9USV0 A0A1B0BCT9 A0A1A9WDQ6 D3TMK2 A0A1S3JM06 T1KKJ9 A0A1L8E4W7 A0A151IEJ7 A0A195EFX4 F4WQW1 A0A158NTJ8 A0A151X3C5 A0A1J1HH80 A0A0P6IHI2 A0A0K8TPP1 B4NQ65 B3MQD0 A0A1W4VTT8 B4R7S5 A0A3B0JXC3 A0A0R3P4F3 B4H4Q6 B4PZD5 B3NVS8 Q29HY3 A0A1Q3G3E7 T1DIT8 Q16YG0 M9NFF8 P40793 H9LJ43 K1PJ53 A0A2R5LJX9 A0A1Z5L6Q1 T1E8E5 A0A154P356
A0A194PL43 A0A212EQJ9 E0VSX8 A0A0L7RAR9 R4WRF3 A0A067R1U4 A0A310SD05 A0A165VG29 A0A2A3E1I2 V5GKL4 N6TXA1 D6WN12 A0A2J7PTR6 A0A1B6GPR3 A0A1B6KK31 A0A1B6J2Q1 A0A1B6FTP5 A0A0K8VS38 A0A023GFP5 G3MKJ3 A0A1E1XJY3 A0A023FF98 L7M3T4 A0A023FUE4 A0A1E1WZ55 A0A224YT61 A0A131Z5G9 A0A131XMD5 W8AM21 A0A034WPG8 A0A0A1WMH4 A0A1W4W8W6 B7PHY7 A0A1B6LTH8 A0A154PP51 A0A0B4J2U1 A0A1S3CZK2 A0A195BRR0 A0A182XWZ5 A0A2M3ZGJ9 A0A2M4A9C9 A0A1Y1KT91 A0A182MGD6 A0A182QIQ5 A0A182VG73 W5JCL2 A0A182NFH5 A0A182K8T7 A0A182UJ57 A0A084VU61 A0A182J293 A0A182LF64 A0A182XI45 F5HK45 A0A182R9Y3 A0A182HT27 Q17031 A0A1A9YJ86 A0A1A9USV0 A0A1B0BCT9 A0A1A9WDQ6 D3TMK2 A0A1S3JM06 T1KKJ9 A0A1L8E4W7 A0A151IEJ7 A0A195EFX4 F4WQW1 A0A158NTJ8 A0A151X3C5 A0A1J1HH80 A0A0P6IHI2 A0A0K8TPP1 B4NQ65 B3MQD0 A0A1W4VTT8 B4R7S5 A0A3B0JXC3 A0A0R3P4F3 B4H4Q6 B4PZD5 B3NVS8 Q29HY3 A0A1Q3G3E7 T1DIT8 Q16YG0 M9NFF8 P40793 H9LJ43 K1PJ53 A0A2R5LJX9 A0A1Z5L6Q1 T1E8E5 A0A154P356
Pubmed
19121390
24866850
26354079
22118469
20566863
23691247
+ More
24845553 23537049 18362917 19820115 22216098 29209593 25576852 28503490 28797301 26830274 28049606 24495485 25348373 25830018 20075255 25244985 28004739 20920257 23761445 24438588 20966253 12364791 14747013 17210077 8917545 20353571 21719571 21347285 26369729 17994087 18057021 15632085 17550304 24330624 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 7958857 10772800 12537569 9371783 12490550 26801180 22484279 22992520 28528879
24845553 23537049 18362917 19820115 22216098 29209593 25576852 28503490 28797301 26830274 28049606 24495485 25348373 25830018 20075255 25244985 28004739 20920257 23761445 24438588 20966253 12364791 14747013 17210077 8917545 20353571 21719571 21347285 26369729 17994087 18057021 15632085 17550304 24330624 17510324 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 7958857 10772800 12537569 9371783 12490550 26801180 22484279 22992520 28528879
EMBL
BABH01016932
NWSH01003565
PCG66103.1
ODYU01009260
SOQ53553.1
KJ547700
+ More
AHZ89304.1 GDQN01008341 GDQN01005206 JAT82713.1 JAT85848.1 KQ461108 KPJ09033.1 KQ459605 KPI91825.1 AGBW02013269 OWR43731.1 DS235758 EEB16484.1 KQ414617 KOC68017.1 AK417262 BAN20477.1 KK853090 KDR11574.1 KQ776822 OAD52173.1 KT984806 AMZ00358.1 KZ288496 PBC25182.1 GALX01003847 JAB64619.1 APGK01051402 KB741190 KB632094 ENN73011.1 ERL88706.1 KQ971343 EFA03253.1 NEVH01021221 PNF19725.1 GECZ01005343 JAS64426.1 GEBQ01028192 JAT11785.1 GECU01032386 GECU01014264 JAS75320.1 JAS93442.1 GECZ01016189 JAS53580.1 GDHF01027837 GDHF01010608 JAI24477.1 JAI41706.1 GBBM01002691 JAC32727.1 JO842394 AEO34011.1 GFAA01004113 JAT99321.1 GBBK01003861 JAC20621.1 GACK01006048 JAA58986.1 GBBL01002028 JAC25292.1 GFAC01006883 JAT92305.1 GFPF01006625 MAA17771.1 GEDV01001990 JAP86567.1 GEFH01000362 JAP68219.1 GAMC01019593 JAB86962.1 GAKP01003309 JAC55643.1 GBXI01014669 JAC99622.1 ABJB010057131 DS715571 EEC06209.1 GEBQ01013003 JAT26974.1 KQ434977 KZC12950.1 AAZX01008259 KQ976424 KYM88626.1 GGFM01006923 MBW27674.1 GGFK01004085 MBW37406.1 GEZM01074314 GEZM01074313 JAV64603.1 AXCM01006797 AXCN02000213 ADMH02001552 GGFL01004220 ETN62087.1 MBW68398.1 ATLV01016645 KE525099 KFB41505.1 AXCP01001391 AAAB01008859 EGK96886.1 EGK96887.1 APCN01000403 Z69980 JXJN01012114 CCAG010004713 EZ422654 ADD18930.1 CAEY01000176 GFDF01000311 JAV13773.1 KQ977870 KYM99231.1 KQ978983 KYN26794.1 GL888275 EGI63474.1 ADTU01025983 KQ982562 KYQ54891.1 CVRI01000002 CRK86914.1 GDIQ01004330 JAN90407.1 GDAI01001510 JAI16093.1 CH964291 EDW86290.1 CH902621 EDV44556.1 KPU81636.1 CM000366 EDX18408.1 OUUW01000011 SPP86715.1 CH379064 KRT06405.1 CH479209 EDW32668.1 CM000162 EDX02091.1 CH954180 EDV46743.1 EAL31624.1 GFDL01000720 JAV34325.1 GALA01000766 JAA94086.1 CH477516 EAT39696.1 AE014298 AFH07474.1 AFH07475.1 U11824 AF153423 AF153424 AF153425 AF153426 AF153427 AF153428 AF153429 AY119570 JF919360 AEF33422.1 JH818665 JH816127 EKC18954.1 EKC28126.1 GGLE01005727 MBY09853.1 GFJQ02003877 JAW03093.1 GAMD01002247 JAA99343.1 KQ434809 KZC06369.1
AHZ89304.1 GDQN01008341 GDQN01005206 JAT82713.1 JAT85848.1 KQ461108 KPJ09033.1 KQ459605 KPI91825.1 AGBW02013269 OWR43731.1 DS235758 EEB16484.1 KQ414617 KOC68017.1 AK417262 BAN20477.1 KK853090 KDR11574.1 KQ776822 OAD52173.1 KT984806 AMZ00358.1 KZ288496 PBC25182.1 GALX01003847 JAB64619.1 APGK01051402 KB741190 KB632094 ENN73011.1 ERL88706.1 KQ971343 EFA03253.1 NEVH01021221 PNF19725.1 GECZ01005343 JAS64426.1 GEBQ01028192 JAT11785.1 GECU01032386 GECU01014264 JAS75320.1 JAS93442.1 GECZ01016189 JAS53580.1 GDHF01027837 GDHF01010608 JAI24477.1 JAI41706.1 GBBM01002691 JAC32727.1 JO842394 AEO34011.1 GFAA01004113 JAT99321.1 GBBK01003861 JAC20621.1 GACK01006048 JAA58986.1 GBBL01002028 JAC25292.1 GFAC01006883 JAT92305.1 GFPF01006625 MAA17771.1 GEDV01001990 JAP86567.1 GEFH01000362 JAP68219.1 GAMC01019593 JAB86962.1 GAKP01003309 JAC55643.1 GBXI01014669 JAC99622.1 ABJB010057131 DS715571 EEC06209.1 GEBQ01013003 JAT26974.1 KQ434977 KZC12950.1 AAZX01008259 KQ976424 KYM88626.1 GGFM01006923 MBW27674.1 GGFK01004085 MBW37406.1 GEZM01074314 GEZM01074313 JAV64603.1 AXCM01006797 AXCN02000213 ADMH02001552 GGFL01004220 ETN62087.1 MBW68398.1 ATLV01016645 KE525099 KFB41505.1 AXCP01001391 AAAB01008859 EGK96886.1 EGK96887.1 APCN01000403 Z69980 JXJN01012114 CCAG010004713 EZ422654 ADD18930.1 CAEY01000176 GFDF01000311 JAV13773.1 KQ977870 KYM99231.1 KQ978983 KYN26794.1 GL888275 EGI63474.1 ADTU01025983 KQ982562 KYQ54891.1 CVRI01000002 CRK86914.1 GDIQ01004330 JAN90407.1 GDAI01001510 JAI16093.1 CH964291 EDW86290.1 CH902621 EDV44556.1 KPU81636.1 CM000366 EDX18408.1 OUUW01000011 SPP86715.1 CH379064 KRT06405.1 CH479209 EDW32668.1 CM000162 EDX02091.1 CH954180 EDV46743.1 EAL31624.1 GFDL01000720 JAV34325.1 GALA01000766 JAA94086.1 CH477516 EAT39696.1 AE014298 AFH07474.1 AFH07475.1 U11824 AF153423 AF153424 AF153425 AF153426 AF153427 AF153428 AF153429 AY119570 JF919360 AEF33422.1 JH818665 JH816127 EKC18954.1 EKC28126.1 GGLE01005727 MBY09853.1 GFJQ02003877 JAW03093.1 GAMD01002247 JAA99343.1 KQ434809 KZC06369.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000009046
+ More
UP000053825 UP000027135 UP000242457 UP000019118 UP000030742 UP000007266 UP000235965 UP000192223 UP000001555 UP000076502 UP000002358 UP000079169 UP000078540 UP000076408 UP000075883 UP000075886 UP000075903 UP000000673 UP000075884 UP000075881 UP000075902 UP000030765 UP000075880 UP000075882 UP000076407 UP000007062 UP000075900 UP000075840 UP000092443 UP000078200 UP000092460 UP000091820 UP000092444 UP000085678 UP000015104 UP000078542 UP000078492 UP000007755 UP000005205 UP000075809 UP000183832 UP000007798 UP000007801 UP000192221 UP000000304 UP000268350 UP000001819 UP000008744 UP000002282 UP000008711 UP000008820 UP000000803 UP000005408
UP000053825 UP000027135 UP000242457 UP000019118 UP000030742 UP000007266 UP000235965 UP000192223 UP000001555 UP000076502 UP000002358 UP000079169 UP000078540 UP000076408 UP000075883 UP000075886 UP000075903 UP000000673 UP000075884 UP000075881 UP000075902 UP000030765 UP000075880 UP000075882 UP000076407 UP000007062 UP000075900 UP000075840 UP000092443 UP000078200 UP000092460 UP000091820 UP000092444 UP000085678 UP000015104 UP000078542 UP000078492 UP000007755 UP000005205 UP000075809 UP000183832 UP000007798 UP000007801 UP000192221 UP000000304 UP000268350 UP000001819 UP000008744 UP000002282 UP000008711 UP000008820 UP000000803 UP000005408
Pfam
PF00071 Ras
Interpro
SUPFAM
SSF52540
SSF52540
CDD
ProteinModelPortal
H9JRY5
A0A2A4J3X5
A0A2H1WKE4
A0A059VBH0
A0A1E1W6W5
A0A194QU25
+ More
A0A194PL43 A0A212EQJ9 E0VSX8 A0A0L7RAR9 R4WRF3 A0A067R1U4 A0A310SD05 A0A165VG29 A0A2A3E1I2 V5GKL4 N6TXA1 D6WN12 A0A2J7PTR6 A0A1B6GPR3 A0A1B6KK31 A0A1B6J2Q1 A0A1B6FTP5 A0A0K8VS38 A0A023GFP5 G3MKJ3 A0A1E1XJY3 A0A023FF98 L7M3T4 A0A023FUE4 A0A1E1WZ55 A0A224YT61 A0A131Z5G9 A0A131XMD5 W8AM21 A0A034WPG8 A0A0A1WMH4 A0A1W4W8W6 B7PHY7 A0A1B6LTH8 A0A154PP51 A0A0B4J2U1 A0A1S3CZK2 A0A195BRR0 A0A182XWZ5 A0A2M3ZGJ9 A0A2M4A9C9 A0A1Y1KT91 A0A182MGD6 A0A182QIQ5 A0A182VG73 W5JCL2 A0A182NFH5 A0A182K8T7 A0A182UJ57 A0A084VU61 A0A182J293 A0A182LF64 A0A182XI45 F5HK45 A0A182R9Y3 A0A182HT27 Q17031 A0A1A9YJ86 A0A1A9USV0 A0A1B0BCT9 A0A1A9WDQ6 D3TMK2 A0A1S3JM06 T1KKJ9 A0A1L8E4W7 A0A151IEJ7 A0A195EFX4 F4WQW1 A0A158NTJ8 A0A151X3C5 A0A1J1HH80 A0A0P6IHI2 A0A0K8TPP1 B4NQ65 B3MQD0 A0A1W4VTT8 B4R7S5 A0A3B0JXC3 A0A0R3P4F3 B4H4Q6 B4PZD5 B3NVS8 Q29HY3 A0A1Q3G3E7 T1DIT8 Q16YG0 M9NFF8 P40793 H9LJ43 K1PJ53 A0A2R5LJX9 A0A1Z5L6Q1 T1E8E5 A0A154P356
A0A194PL43 A0A212EQJ9 E0VSX8 A0A0L7RAR9 R4WRF3 A0A067R1U4 A0A310SD05 A0A165VG29 A0A2A3E1I2 V5GKL4 N6TXA1 D6WN12 A0A2J7PTR6 A0A1B6GPR3 A0A1B6KK31 A0A1B6J2Q1 A0A1B6FTP5 A0A0K8VS38 A0A023GFP5 G3MKJ3 A0A1E1XJY3 A0A023FF98 L7M3T4 A0A023FUE4 A0A1E1WZ55 A0A224YT61 A0A131Z5G9 A0A131XMD5 W8AM21 A0A034WPG8 A0A0A1WMH4 A0A1W4W8W6 B7PHY7 A0A1B6LTH8 A0A154PP51 A0A0B4J2U1 A0A1S3CZK2 A0A195BRR0 A0A182XWZ5 A0A2M3ZGJ9 A0A2M4A9C9 A0A1Y1KT91 A0A182MGD6 A0A182QIQ5 A0A182VG73 W5JCL2 A0A182NFH5 A0A182K8T7 A0A182UJ57 A0A084VU61 A0A182J293 A0A182LF64 A0A182XI45 F5HK45 A0A182R9Y3 A0A182HT27 Q17031 A0A1A9YJ86 A0A1A9USV0 A0A1B0BCT9 A0A1A9WDQ6 D3TMK2 A0A1S3JM06 T1KKJ9 A0A1L8E4W7 A0A151IEJ7 A0A195EFX4 F4WQW1 A0A158NTJ8 A0A151X3C5 A0A1J1HH80 A0A0P6IHI2 A0A0K8TPP1 B4NQ65 B3MQD0 A0A1W4VTT8 B4R7S5 A0A3B0JXC3 A0A0R3P4F3 B4H4Q6 B4PZD5 B3NVS8 Q29HY3 A0A1Q3G3E7 T1DIT8 Q16YG0 M9NFF8 P40793 H9LJ43 K1PJ53 A0A2R5LJX9 A0A1Z5L6Q1 T1E8E5 A0A154P356
PDB
5C2J
E-value=4.65441e-87,
Score=815
Ontologies
PATHWAY
GO
GO:0005525
GO:0003924
GO:0007264
GO:0042995
GO:0008360
GO:0005886
GO:0007015
GO:0030031
GO:0005938
GO:0007163
GO:0032488
GO:0007266
GO:0006897
GO:0032956
GO:0005737
GO:0030036
GO:0019901
GO:0051301
GO:0004767
GO:0005912
GO:0007275
GO:0007286
GO:0030011
GO:0008045
GO:0030866
GO:0048813
GO:0051601
GO:0016318
GO:0007274
GO:0048675
GO:0045177
GO:0060446
GO:0046330
GO:0045200
GO:0005634
GO:0035099
GO:0050770
GO:0045185
GO:0030833
GO:0090303
GO:0010592
GO:0030041
GO:0030027
GO:0035011
GO:0007391
GO:0032794
GO:0097206
GO:0031532
GO:0050975
GO:0008582
GO:0042060
GO:0035318
GO:0046843
GO:0016028
GO:0006909
GO:0045860
GO:0030426
GO:0051491
GO:0048010
GO:0072659
GO:0007411
GO:0030707
GO:0048477
GO:0007349
GO:0007409
GO:0009611
GO:0030424
GO:0048812
GO:0007179
GO:0031175
GO:0016192
GO:0035010
GO:0005515
GO:0008270
GO:0030126
GO:0005488
GO:0003677
GO:0003707
GO:0007169
GO:0016021
Topology
Subcellular location
Cell junction
Adherens junction
Cell membrane
Adherens junction
Cell membrane
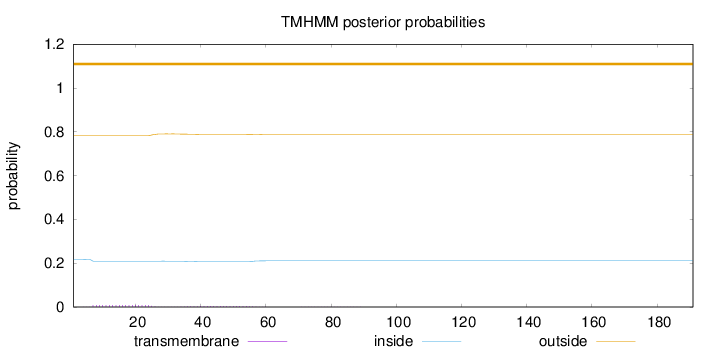
Length:
191
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.25725
Exp number, first 60 AAs:
0.25301
Total prob of N-in:
0.21798
outside
1 - 191
Population Genetic Test Statistics
Pi
15.58678
Theta
16.965683
Tajima's D
-1.146934
CLR
0.417047
CSRT
0.107444627768612
Interpretation
Uncertain
