Gene
KWMTBOMO00476
Pre Gene Modal
BGIBMGA012297
Annotation
PREDICTED:_sterol_regulatory_element-binding_protein_cleavage-activating_protein_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 3.688
Sequence
CDS
ATGGCAGGATCTACGCTTCCCGAGAGGGTAGCTCAAATTTATTATTCATATGGATTATTTTGTTCGTCGTACCCAATAACTGCAATCACGCTCGCACTCTCCATCGTACTGTTATGCTGTTATCCTCTTCTGAATGTTCCACTACCAGGAAATGTACCTACAGCGATCAATTTTCCACTTAACAGTGACCATCTTAGACAATCAACAGATCCTGGATGTGCAGATAATAAATGTACATCGACACCTACTTCTCAGTTGGACCTTCTATTCAATGAAACGCAAAGACTTCCATATATTTGGGCAAAGGACAAACCGCTACTGTATGTTCAACAAATCATTTTAAAAATTGGCGTATCACCATGGAATTCGAATTTGAAAATGTGGGACGCGTTCCGGGCTCCAATCCAAGAAGTGTTCCGGTTGTTAGAAACCATCAGAAATCACGAAGATCCAGATTTAAAACAAACTCTACAACAGAACTGCTATCAAATCGGAGCTATCAAACGACGCGAAAGCAAAGCCAGCATCGAACGTGTCCTACCGGAATACAGTTGTCTCCTTCTGTCACCAGCGAATTTGTGGCAACAAAACCTTCAAACGTTTTCACTCGACACTAACATAGTCAACACCGTTTACAACTATCAGAACTTGCAAAAAGGGAAAGTTTCAATAGCGGAAATGGCGTTCGGCATGCAAATGCGGGACACCGGAATAAAGCGGTATCCGTTGCGGGCCCGACCTCGTGTACTGCAATACGCTGTCACAATTTTCTACGACAACCCCGAAGACAGATTTATTCAATCACTCACCAAGAAACTCCAAGATATGTACCCCCTATATCAAGACAAGTCGCACGTCAATGTTGACGAGGTGACTCTCGTATATTATCCGGGGCAGTTCAATTACCGGGAACTCGTCCCATTGACAATAACGTTTGTCGGTTTATTTCTCTACGTATATTTTTCTGTAAGGAAGATCGAGTACATAAGATCTAAATTAGGACTCGCGGCCTGCGCCGTCGTGACGATAGCCGGGAGTTTAAGTATGTCGATGGGGATTTGTTTTTACTTCGGCTTCTCGTTGAGTCACCAGGGAAAAGAAATATTTCCGTATCTCATCATCATCGTTGGCTTGGAAAACGTATTAGTGTTGACCAAGAGCGTAACTTCGACCGACTCGAGGCTGGATGTCAAGATACGATTGGCGCAGGGACTCAGCAGGGAGGGATGGTCGATCACCAAAAACTTACTAACCGAAATCACTATATTCACAGTCAGTTTCTTTACTTTCGTACCGTTCATACAGGAGTTCTCCATATTCATGATAGTCAGTTTGTTGTCCGATTATTTTATACAAATTTTGTTTTTCGCTACCGTTCTCGGTATCGACGTGCGTCACATCGAGTACCTCCAAGATAAAAATAATAAATTGCACTTAAAAGAGTATTTTAGTATGCACAACGCGCTCAACTGGCGTTACAATAAAAGTTACACTGACGAGTTTAACTTTTCGCCGCAACCGATGACGAAATCGAAGTCCCATCCGAGACTGATCGGATTGGCCCAGAACAGCCCCACGGACGTGGTCGCCAACCGGAACTCTAGTCCGGGACATCAGGACAATCGCGTGCCGAAACGCATCAAGCTTGTGAACATGTGGGCGAGGACTAGGTTTTTTCAACGGGCCTTCATGCTGTGGATGCTCGTGTGGATATCCACGATCGTCTACGATTCCGGTGTCGTCGACTATTTCATTGGGAGCGCGGAAAAATTAGAAAACGTCGAGACGAACAAAAATTACGTCGATAAAAGGGAATCTACAAAGGAATCAATGTACGAGATGTTTTATAATAGTACAGATCGAAATACGGTGGTGTTCCCGCCCATGTTCGATATGGCCGACTCTGATAAAGCACTCGACACAAACGACACTATAATGTTGAAGCATTCTCCGCATTCGCGGCCGTGGTCTAGGCTTTCTGACTACCATTGGTCAACTATACTATCCAACTACAATGAATCGACAGCGGGTCGTTACGTTGTAATGCTACCACCAACGCTACTGAGTCATCGCGTAAGTCCCGAACTTGCTGTGGGAGTTCGGAATCCGGACGAGAAGGATCCACCACCGCTGAGATGGCAAGCATTAGCTGCAGCGCTTGATCCCATTGACATACTACCCGATTTCGACGTTGTCGACGGGAAAGGCTCCCCGCTCCAATGGGGCAAGGGGTCCGAACTGCCTATATACCCTACAACACCAATGGAAATTCTTCTGCTCGCCATACTGTGCTCCATAAGCGTCGCCGTCATCGCCTACATGATGGTGGTTTTGTACAGGTGTATTTGTTCACGGCATTATGCGGAATGGAGGGCTTCTTGGAACGATAACGAGGAATACCGGAGGATCATAGCCAAGCAACCGGCCATTCAGTTGGTAATGGAAGCAGTGCCTTTGGTCGTAGCCGGCCATAGCCAGGAAGTGGAGTGCCTGGTGACCGATGGTGAGAAGATAGTTAGTTCTTGCCTCCAAGGCAATATAAAAGTATGGGACTCGCAGAACGGAGAAATCATCACAAACATTGATAGAGAAGCTTTCTTCAACCTACAGAATCTATTACACAGTAAAGCTAAATCAAAAAGGTGCAGTTATCACGGTGACCTACGTGATAATGAAGATGCCTACGAAGTAGTTCCAACATCGAAAAATAACAAGCCAGTTCCTCAATTGCGAAAAGGTTTGAGTTCGTATTTAAGTGGATTGCGATTTCGTCCCGCATCGAGCGAGCGTGTGCCGAATCTTTCCGTAGCGGACACCACTAAGTATGAATTCGCGAAAAGTTACAGATCTATATACTACACAAACGAAAATGACGACGTTTATAATTGTAACGAATGCAAAGAAAACATTGACATAAACAATTTAAACATAGCCAAAACTAACAAAAGTACTAACAGTATTCAAGAAGATGACTTAAGTAAAGATATATTAGAGAATATGGAAGCTAGCAATAATCTAAGAAATAGAAGTAAGAATAAAACGTATAATGAGGAGCGACCGAGAACTAAGCAAGCAAGCGGTTCAGCTGCTACCAGTACCTTGGATTGGCAAGGGAAACCGATTCTAGAATCTCCTGTCTGGTGTATGGACTTTAGCAATGATCTCATCATATTAGGCTGTGCGGATGGCAGACTTGAGTTCTGGGAAGCGAGCACGGGACGACTTATGTGCGTGTGCGGGGGCGGCGAGGGCGTGACGCACGTGCGGGCGCTGGCGGGCGCGGCGCGGGTGTGTGCGGCCTCGCTCTCCGGGCACCTCACTCTGATGCGCCTCGACGCCTACAACAGTGCGTCGGGCGCCCATGTCGACTGGGGCTTCAGCACCGCGCACAGACGAACGCACAAGCGGAACGGTTCGACCGGCTCGTTACAGACGAGCTTCGGCGTGAACGACGAAAGCTCCCGATACAGAACTAATTTCACCTACAAAAGCGATCCTAATGACGGAGAAGAGGTGGTGTGCGTTCGAATAGCGCAGTGCAGGCCTCATCAGCAGCCTGTTACTGAGATGCACTCGGAAGGCGGTCGCATCTTAACCGGAGGTCAGGATCACGTGGTCAAGGTGTTCTCTAGCGCGGAATTAACGCCTCTCTTTACACTGCACGGACACTGCGGACCTATAACCAGTTGCTTCATCGATCACGCGACTCCAACGATAGCTGGGAGCGGATCGCAGGACGGCCTCCTCTGCGTGTGGGACCTGCATACTGGCGCGTGCCTGTACAGTATGCAGGCTCACGACGGCGCAGTGACGTCACTGGCGTACACCGCCTCGTACGTTGTGTCGGCGGGCGCTGATGAACGACTCTGCATATGGGATCGCTTTCAAGGGCATATGCTCAACTCCATACACGTGGGGCTCAATTATATGAGTCGTATGCTGCCCTTAACTCACACCCTGCTCGTGATGGGCGACAGAAGCGGCATCACCGTCTACGACCTGAGCAGCGGTGACGTCATAAGACGAGTTTTACTCGGTCAAAGCGACGGGTGTATTTTCGTAAGACAAATATTGCCTCTGAAAGACGCTATTGTGTGCGATTACGCGAATCAATTGAGGATAGTGCGCTTCCCTCTCGTCTCTCGGCTCGATTTAAAGAGCGACTAA
Protein
MAGSTLPERVAQIYYSYGLFCSSYPITAITLALSIVLLCCYPLLNVPLPGNVPTAINFPLNSDHLRQSTDPGCADNKCTSTPTSQLDLLFNETQRLPYIWAKDKPLLYVQQIILKIGVSPWNSNLKMWDAFRAPIQEVFRLLETIRNHEDPDLKQTLQQNCYQIGAIKRRESKASIERVLPEYSCLLLSPANLWQQNLQTFSLDTNIVNTVYNYQNLQKGKVSIAEMAFGMQMRDTGIKRYPLRARPRVLQYAVTIFYDNPEDRFIQSLTKKLQDMYPLYQDKSHVNVDEVTLVYYPGQFNYRELVPLTITFVGLFLYVYFSVRKIEYIRSKLGLAACAVVTIAGSLSMSMGICFYFGFSLSHQGKEIFPYLIIIVGLENVLVLTKSVTSTDSRLDVKIRLAQGLSREGWSITKNLLTEITIFTVSFFTFVPFIQEFSIFMIVSLLSDYFIQILFFATVLGIDVRHIEYLQDKNNKLHLKEYFSMHNALNWRYNKSYTDEFNFSPQPMTKSKSHPRLIGLAQNSPTDVVANRNSSPGHQDNRVPKRIKLVNMWARTRFFQRAFMLWMLVWISTIVYDSGVVDYFIGSAEKLENVETNKNYVDKRESTKESMYEMFYNSTDRNTVVFPPMFDMADSDKALDTNDTIMLKHSPHSRPWSRLSDYHWSTILSNYNESTAGRYVVMLPPTLLSHRVSPELAVGVRNPDEKDPPPLRWQALAAALDPIDILPDFDVVDGKGSPLQWGKGSELPIYPTTPMEILLLAILCSISVAVIAYMMVVLYRCICSRHYAEWRASWNDNEEYRRIIAKQPAIQLVMEAVPLVVAGHSQEVECLVTDGEKIVSSCLQGNIKVWDSQNGEIITNIDREAFFNLQNLLHSKAKSKRCSYHGDLRDNEDAYEVVPTSKNNKPVPQLRKGLSSYLSGLRFRPASSERVPNLSVADTTKYEFAKSYRSIYYTNENDDVYNCNECKENIDINNLNIAKTNKSTNSIQEDDLSKDILENMEASNNLRNRSKNKTYNEERPRTKQASGSAATSTLDWQGKPILESPVWCMDFSNDLIILGCADGRLEFWEASTGRLMCVCGGGEGVTHVRALAGAARVCAASLSGHLTLMRLDAYNSASGAHVDWGFSTAHRRTHKRNGSTGSLQTSFGVNDESSRYRTNFTYKSDPNDGEEVVCVRIAQCRPHQQPVTEMHSEGGRILTGGQDHVVKVFSSAELTPLFTLHGHCGPITSCFIDHATPTIAGSGSQDGLLCVWDLHTGACLYSMQAHDGAVTSLAYTASYVVSAGADERLCIWDRFQGHMLNSIHVGLNYMSRMLPLTHTLLVMGDRSGITVYDLSSGDVIRRVLLGQSDGCIFVRQILPLKDAIVCDYANQLRIVRFPLVSRLDLKSD
Summary
Uniprot
H9JRY7
A0A2H1WVZ6
A0A194QZL4
A0A194PEN5
A0A2A4J5H9
A0A212EQI2
+ More
A0A2A4J3T2 A0A2A4J5E1 A0A232EU08 A0A310SJD1 A0A195C2A4 A0A0L7RAN3 A0A088ASY4 A0A158NN36 A0A151JQA8 A0A195BTS1 A0A1Y1M6P5 E2AJT3 F4X7I1 A0A0C9QWW6 A0A1W4WPE7 A0A3L8D4W0 D6WLE2 A0A0J7NKP0 E2BGB3 A0A026W3S0 W4VSE5 A0A182RLJ0 A0A182MRB0 A0A1B6CPD3 A0A1Q3F4H9 A0A1B6DX23 A0A1Q3F4L8 A0A1Q3F4Q8 A0A2A3E702 A0A084WCI9 A0A336L177 A0A182P5P6 A0A182LH98 A0A182JU38 E0VRD4 A0A1B6GIX4 A0A182UP68 A0A182HDQ9 A0A336MGS4 A0A1J1HRT4 B0X214 A0A182HY49 A0A182WXH4 A0A1D2MRY1 A0A0A9YBG3 A0A2S2R3Z4 J9K836 A0A2H8TLV4 A0A1B6I5R4 A0A182TP90 A0A0K8U0I1 L7MHT6 A0A131YWP8
A0A2A4J3T2 A0A2A4J5E1 A0A232EU08 A0A310SJD1 A0A195C2A4 A0A0L7RAN3 A0A088ASY4 A0A158NN36 A0A151JQA8 A0A195BTS1 A0A1Y1M6P5 E2AJT3 F4X7I1 A0A0C9QWW6 A0A1W4WPE7 A0A3L8D4W0 D6WLE2 A0A0J7NKP0 E2BGB3 A0A026W3S0 W4VSE5 A0A182RLJ0 A0A182MRB0 A0A1B6CPD3 A0A1Q3F4H9 A0A1B6DX23 A0A1Q3F4L8 A0A1Q3F4Q8 A0A2A3E702 A0A084WCI9 A0A336L177 A0A182P5P6 A0A182LH98 A0A182JU38 E0VRD4 A0A1B6GIX4 A0A182UP68 A0A182HDQ9 A0A336MGS4 A0A1J1HRT4 B0X214 A0A182HY49 A0A182WXH4 A0A1D2MRY1 A0A0A9YBG3 A0A2S2R3Z4 J9K836 A0A2H8TLV4 A0A1B6I5R4 A0A182TP90 A0A0K8U0I1 L7MHT6 A0A131YWP8
Pubmed
EMBL
BABH01016927
BABH01016928
ODYU01011417
SOQ57146.1
KQ461108
KPJ09036.1
+ More
KQ459605 KPI91821.1 NWSH01003266 PCG66652.1 AGBW02013269 OWR43735.1 PCG66651.1 PCG66650.1 NNAY01002188 OXU21849.1 KQ764421 OAD54522.1 KQ978344 KYM94957.1 KQ414618 KOC67890.1 ADTU01020975 KQ978705 KYN29132.1 KQ976406 KYM91676.1 GEZM01040958 JAV80688.1 GL440100 EFN66249.1 GL888840 EGI57619.1 GBYB01005167 JAG74934.1 QOIP01000013 RLU15289.1 KQ971343 EFA03461.2 LBMM01003878 KMQ93030.1 GL448138 EFN85276.1 KK107467 EZA50256.1 GANO01000062 JAB59809.1 AXCM01001256 GEDC01021998 JAS15300.1 GFDL01012574 JAV22471.1 GEDC01007082 JAS30216.1 GFDL01012617 JAV22428.1 GFDL01012532 JAV22513.1 KZ288361 PBC27006.1 ATLV01022672 ATLV01022673 ATLV01022674 KE525335 KFB47933.1 UFQS01001435 UFQT01001435 SSX10880.1 SSX30558.1 DS235465 EEB15940.1 GECZ01007398 JAS62371.1 JXUM01130316 KQ567562 KXJ69369.1 UFQT01001190 SSX29395.1 CVRI01000020 CRK90733.1 DS232276 EDS38944.1 APCN01004491 LJIJ01000613 ODM95859.1 GBHO01013183 GBHO01013182 GDHC01021140 JAG30421.1 JAG30422.1 JAP97488.1 GGMS01015523 MBY84726.1 ABLF02037163 GFXV01003175 MBW14980.1 GECU01025466 JAS82240.1 GDHF01032494 JAI19820.1 GACK01001273 JAA63761.1 GEDV01006216 JAP82341.1
KQ459605 KPI91821.1 NWSH01003266 PCG66652.1 AGBW02013269 OWR43735.1 PCG66651.1 PCG66650.1 NNAY01002188 OXU21849.1 KQ764421 OAD54522.1 KQ978344 KYM94957.1 KQ414618 KOC67890.1 ADTU01020975 KQ978705 KYN29132.1 KQ976406 KYM91676.1 GEZM01040958 JAV80688.1 GL440100 EFN66249.1 GL888840 EGI57619.1 GBYB01005167 JAG74934.1 QOIP01000013 RLU15289.1 KQ971343 EFA03461.2 LBMM01003878 KMQ93030.1 GL448138 EFN85276.1 KK107467 EZA50256.1 GANO01000062 JAB59809.1 AXCM01001256 GEDC01021998 JAS15300.1 GFDL01012574 JAV22471.1 GEDC01007082 JAS30216.1 GFDL01012617 JAV22428.1 GFDL01012532 JAV22513.1 KZ288361 PBC27006.1 ATLV01022672 ATLV01022673 ATLV01022674 KE525335 KFB47933.1 UFQS01001435 UFQT01001435 SSX10880.1 SSX30558.1 DS235465 EEB15940.1 GECZ01007398 JAS62371.1 JXUM01130316 KQ567562 KXJ69369.1 UFQT01001190 SSX29395.1 CVRI01000020 CRK90733.1 DS232276 EDS38944.1 APCN01004491 LJIJ01000613 ODM95859.1 GBHO01013183 GBHO01013182 GDHC01021140 JAG30421.1 JAG30422.1 JAP97488.1 GGMS01015523 MBY84726.1 ABLF02037163 GFXV01003175 MBW14980.1 GECU01025466 JAS82240.1 GDHF01032494 JAI19820.1 GACK01001273 JAA63761.1 GEDV01006216 JAP82341.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000218220
UP000007151
UP000215335
+ More
UP000078542 UP000053825 UP000005203 UP000005205 UP000078492 UP000078540 UP000000311 UP000007755 UP000192223 UP000279307 UP000007266 UP000036403 UP000008237 UP000053097 UP000075900 UP000075883 UP000242457 UP000030765 UP000075885 UP000075882 UP000075881 UP000009046 UP000075903 UP000069940 UP000249989 UP000183832 UP000002320 UP000075840 UP000076407 UP000094527 UP000007819 UP000075902
UP000078542 UP000053825 UP000005203 UP000005205 UP000078492 UP000078540 UP000000311 UP000007755 UP000192223 UP000279307 UP000007266 UP000036403 UP000008237 UP000053097 UP000075900 UP000075883 UP000242457 UP000030765 UP000075885 UP000075882 UP000075881 UP000009046 UP000075903 UP000069940 UP000249989 UP000183832 UP000002320 UP000075840 UP000076407 UP000094527 UP000007819 UP000075902
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9JRY7
A0A2H1WVZ6
A0A194QZL4
A0A194PEN5
A0A2A4J5H9
A0A212EQI2
+ More
A0A2A4J3T2 A0A2A4J5E1 A0A232EU08 A0A310SJD1 A0A195C2A4 A0A0L7RAN3 A0A088ASY4 A0A158NN36 A0A151JQA8 A0A195BTS1 A0A1Y1M6P5 E2AJT3 F4X7I1 A0A0C9QWW6 A0A1W4WPE7 A0A3L8D4W0 D6WLE2 A0A0J7NKP0 E2BGB3 A0A026W3S0 W4VSE5 A0A182RLJ0 A0A182MRB0 A0A1B6CPD3 A0A1Q3F4H9 A0A1B6DX23 A0A1Q3F4L8 A0A1Q3F4Q8 A0A2A3E702 A0A084WCI9 A0A336L177 A0A182P5P6 A0A182LH98 A0A182JU38 E0VRD4 A0A1B6GIX4 A0A182UP68 A0A182HDQ9 A0A336MGS4 A0A1J1HRT4 B0X214 A0A182HY49 A0A182WXH4 A0A1D2MRY1 A0A0A9YBG3 A0A2S2R3Z4 J9K836 A0A2H8TLV4 A0A1B6I5R4 A0A182TP90 A0A0K8U0I1 L7MHT6 A0A131YWP8
A0A2A4J3T2 A0A2A4J5E1 A0A232EU08 A0A310SJD1 A0A195C2A4 A0A0L7RAN3 A0A088ASY4 A0A158NN36 A0A151JQA8 A0A195BTS1 A0A1Y1M6P5 E2AJT3 F4X7I1 A0A0C9QWW6 A0A1W4WPE7 A0A3L8D4W0 D6WLE2 A0A0J7NKP0 E2BGB3 A0A026W3S0 W4VSE5 A0A182RLJ0 A0A182MRB0 A0A1B6CPD3 A0A1Q3F4H9 A0A1B6DX23 A0A1Q3F4L8 A0A1Q3F4Q8 A0A2A3E702 A0A084WCI9 A0A336L177 A0A182P5P6 A0A182LH98 A0A182JU38 E0VRD4 A0A1B6GIX4 A0A182UP68 A0A182HDQ9 A0A336MGS4 A0A1J1HRT4 B0X214 A0A182HY49 A0A182WXH4 A0A1D2MRY1 A0A0A9YBG3 A0A2S2R3Z4 J9K836 A0A2H8TLV4 A0A1B6I5R4 A0A182TP90 A0A0K8U0I1 L7MHT6 A0A131YWP8
PDB
6G6N
E-value=8.45317e-14,
Score=192
Ontologies
GO
PANTHER
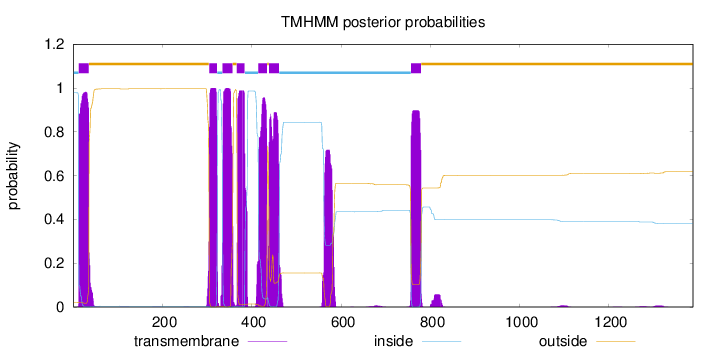
Topology
Length:
1388
Number of predicted TMHs:
7
Exp number of AAs in TMHs:
162.526800000001
Exp number, first 60 AAs:
23.13646
Total prob of N-in:
0.97823
POSSIBLE N-term signal
sequence
inside
1 - 12
TMhelix
13 - 35
outside
36 - 304
TMhelix
305 - 322
inside
323 - 334
TMhelix
335 - 357
outside
358 - 366
TMhelix
367 - 384
inside
385 - 414
TMhelix
415 - 434
outside
435 - 438
TMhelix
439 - 461
inside
462 - 756
TMhelix
757 - 779
outside
780 - 1388
Population Genetic Test Statistics
Pi
20.955282
Theta
22.409524
Tajima's D
-0.968686
CLR
0.027353
CSRT
0.143342832858357
Interpretation
Uncertain
