Gene
KWMTBOMO00472
Pre Gene Modal
BGIBMGA012299
Annotation
PREDICTED:_pleckstrin_homology_domain-containing_family_G_member_5_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 2.647
Sequence
CDS
ATGGCACACACCGAGATCGGCACCCTGTCGGCTACTCATTCTCCAGTTTCTGTGAGAGCGGAGCCTCATGCGAACTTCGAACTATCGGCCCTGCTTACCGAATTCACAAAAAATGGGGTCCCACCTATCAGGGGATTTGCACTCGCCAACATATGCAAAGAAGCCTTCGATTACCTCGTGTGGGCTGAAGAGCCGCCGAATCTGGAACAAGTATACGATTGCAAAAAGCTACCTTCTAATGAGGAAGCAAGGCAGGCTGTGGTCAGGGAACTTGTGAACACAGAAAGTATTTACATCCGTCACCTCACCGCCATCGTAGAGGTATTCATAGCCGCTGCACACGCTCTGCAAGACATGGGTAAACTGCTAGAAGTGGATACTGAGGGGCTTTTCACTAACATCCCTGACGTCCTCAATGCCAGCCTTCACTTCTGGGAGCTTACCTTCTATCCAATGATCATCGATGCAGTCCAGAACAAAGTACCTTTTAACACCGAACTGATGTCGCCCGGCTTCTGCCGTTTCCGTGATTTATTCTACCCCTATGAAAAATACGTCAACACCCAATCGAAAGCATTGGATTATTTTCGCTCTCTATCCAGTAACCCTGATTTTATTGCATACATTTCCTGGTGTCACGCTCATAAATCATGCAACCGGCTGCAACTCTCGGATATAATGGTGAAACCAATGCAGAGACTCACTAAATACGCGCTAATCATTCGTCGTATAATAACACACACTGACACAGAACCGGAACGAACTTCACTCATCGCAATGGAATCATTTGCGAAGAACTACGTGATCGACATAAACCGTACTATTCGGCAGCGGGAGGAGATCGAGAAAATCGAGATGCTGTCAAACATCATTGAATCCTACGAAGTCGAGTTCAAAGACGAGGATCTCGATAGATGCTTTAGGATGTATGCCAATCTGAATCTTAAAGCACCGATGGTCAACTGCATTCCGAGCCAAATGCGAACGTTAATCTACGAAGGTGATCTGCGCTTTAAGGACAGCGTCAAGGAGATCGAAGTCCGGGCCTTTCTGTTGACGGACATGTTACTAATTTGCAAGAAAATCTCCAAAGGGAGTACTTACCAATATAAAATGATCAGACCTAAATTCATGGTTGACAAGATTGTTCCGTTTCCAAAGTATAATCCTCGTACCACTAAGGATTTGCTGTCACTGGTATACGTGGTAGTCGACGAAGTGGGATCGGCTTACAATGCTTTTGCCCTTAGTGAGCCATCAAAAGAATCCGGAGGATCACTTAGACTTTGGGAGCAAAAAGTGCGCGAGGCAAGATTGACTTATGACCTCAGCGTGTGGTACGCCAGAAACCCATCCCGAGATATCTCTGAAATGGAGATGGACTCCTCCTCGGATTACGCCATGAGCGCCACTGGCAAGGGAATAAAACAGAGTTCAGAGGACTTGAACATTGAACGCGAGGCTCGTGAACGAGTGGCAGCCATGCTCCACCGAAACATGGGAGCTTCAACCGAATATGACTTCTCCCAGGCCTCGATGAACACAGACTCTTTCGACGGTATGACAAGCGGCGGTCGCACCACCGGTCTGCATAGCCTGCGCTACCAGATGCACCGAACCTCTACCGGAAGCTCAAGGAACTCACGCCTCTCAAGTTTCCAGCACTCTGTCAGTGCCGCTTCTCATGATGAGCCGCAGCCTGGCACCAGTAAGGCTGGCCTGTTCCTGAAAGCGGGCCCATCCGTGGAACACCCAATTCCAGCATTGACTGTCGAAGATGGAGAATCTCAGGGAGGAGCATCCACGATCACTGTGAACGTGGTCAGCGAGAGCGAATCAGAAACCGTTGTCCAAGGTGATCTCTCTCCCGTGGTTTTACCTGTTCCGGTTCACGTCCCGGGAGTTCAGCCACCACCGCCTGTGGTGGTGCTGCAACACCCTTCGCCGTACAAGCCTCCCATGCGCAGCGTGTCCGCCACCCGCCACACTCTGCGCGTACAACCGAAGTCCGGAATATACGCTCTTGTACACAGCTTACCGGATTTAACAATTGATCAACTGGCCTTAGCCCGCGACCGCCACTCGCACAGCGCCGGACCACCGTCTGCTTCTGAGAAACTGTACCAATCTCACCAAGAATTATTACAAAAAAATCGTCTTTCGACGTCTCAAAACCAATACCTAAGCCCTGACCACCGCAGCGCGAGTCAGCCGCCCTCCAGCCCTACTAGAGCGTCGCTCAAACGAGGACTAGCGTTCTCCTACTCATTCAAGAATCCTCCACTTCACAAAATGGGACATGTAAACAGCCAATCTCAGGTTCAAGCCGAACCCACGCCTTCAACAAGTCAACCATCAAAGCCGGAAATAGAGGCTGTGAGTCCTCAACCAGGCCCAAGTACTAGCACGGATAAAGCCGAGAAGAAAACAAAACAAGTCAGCACCAGTGCATTCTTCACGTCTGGAGGGTGGCACAGCAGGTTCGAAGGATGA
Protein
MAHTEIGTLSATHSPVSVRAEPHANFELSALLTEFTKNGVPPIRGFALANICKEAFDYLVWAEEPPNLEQVYDCKKLPSNEEARQAVVRELVNTESIYIRHLTAIVEVFIAAAHALQDMGKLLEVDTEGLFTNIPDVLNASLHFWELTFYPMIIDAVQNKVPFNTELMSPGFCRFRDLFYPYEKYVNTQSKALDYFRSLSSNPDFIAYISWCHAHKSCNRLQLSDIMVKPMQRLTKYALIIRRIITHTDTEPERTSLIAMESFAKNYVIDINRTIRQREEIEKIEMLSNIIESYEVEFKDEDLDRCFRMYANLNLKAPMVNCIPSQMRTLIYEGDLRFKDSVKEIEVRAFLLTDMLLICKKISKGSTYQYKMIRPKFMVDKIVPFPKYNPRTTKDLLSLVYVVVDEVGSAYNAFALSEPSKESGGSLRLWEQKVREARLTYDLSVWYARNPSRDISEMEMDSSSDYAMSATGKGIKQSSEDLNIEREARERVAAMLHRNMGASTEYDFSQASMNTDSFDGMTSGGRTTGLHSLRYQMHRTSTGSSRNSRLSSFQHSVSAASHDEPQPGTSKAGLFLKAGPSVEHPIPALTVEDGESQGGASTITVNVVSESESETVVQGDLSPVVLPVPVHVPGVQPPPPVVVLQHPSPYKPPMRSVSATRHTLRVQPKSGIYALVHSLPDLTIDQLALARDRHSHSAGPPSASEKLYQSHQELLQKNRLSTSQNQYLSPDHRSASQPPSSPTRASLKRGLAFSYSFKNPPLHKMGHVNSQSQVQAEPTPSTSQPSKPEIEAVSPQPGPSTSTDKAEKKTKQVSTSAFFTSGGWHSRFEG
Summary
Uniprot
EMBL
Proteomes
PRIDE
Pfam
PF00621 RhoGEF
SUPFAM
SSF48065
SSF48065
Gene 3D
CDD
ProteinModelPortal
PDB
5FI1
E-value=1.12992e-11,
Score=172
Ontologies
GO
PANTHER
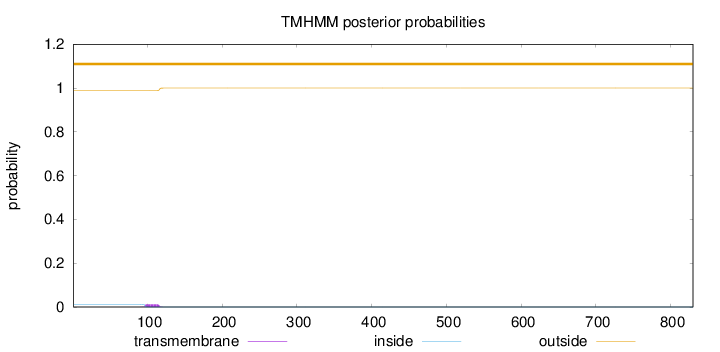
Topology
Length:
830
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.2404
Exp number, first 60 AAs:
0.00021
Total prob of N-in:
0.01223
outside
1 - 830
Population Genetic Test Statistics
Pi
26.77177
Theta
25.955575
Tajima's D
-1.874501
CLR
0.043233
CSRT
0.0220488975551222
Interpretation
Possibly Positive selection
