Gene
KWMTBOMO00466
Pre Gene Modal
BGIBMGA013323
Annotation
PREDICTED:_uncharacterized_protein_LOC101735879_[Bombyx_mori]
Location in the cell
Extracellular Reliability : 1.945 Nuclear Reliability : 1.425
Sequence
CDS
ATGTGTGTCGCTTTCAATAAGCAGAGCCCGAGACTTTATGCGTCCGGTATGACTAATTGGATGAGATATCAGATTTTGCTTTTACACAACCGTCATAGAGACAACATCGCTATGGGCTTGGAAGCTGGTCAACCACCCGCCACCAACATGAGGAAGATGTATTGGGATTACGAACTCGAAGAGATGGCGGAACTGTGGGCGCGTCAATGTCAGAAACGTCACGACGAATGCAGGAATACTATTAGGTTTAATGCACTTCAAAACATGGACGTGCGTCCCGTAGTTCCTAAAGTTACCGAAGCTCAGTTATTGGCTGACGCAGTTGGAGCGTGGTTCTCGGGATCACGGGTGCTACAGTCAGAGCATGTATGGTCGTTCAGGCACGGTAACTGTAGCGCCCCCGGTGGATGCAATTCTCATTGGTATTCAACGGCAGCTTGGGCGTCTACTTGGCGTCTTGGTTGCTCTCAAGCTTTGTGTACTGCCACGAAAAAATCATGCTCGCGTTTTCGCAGGAGAGCAACACCGTCAGAAGACTTGACAAACACATCGACGCGAGTCACCGGCGAGAACATTTTAAGGAGGACCAGGTACATGACGACGGCAACGTTTCCGAAGTTTAAAGATTTCGCGACTACGGTGAGAGTGAAGACGAAGGAATCGCCGCTGCATCGAGGTATTTTGGAGATGACGAACGTACGTAAAGCAAATGATGAACCGAAAATCGTCGCATTCACGTTCTGTAACTACGGCCCGGCCGGGAACATCGATGGGTACCCGATTTACGAAGCCGGTCTGTCATGCAGTAACTGCCCACAGGGGACGCGATGCGGGGATCGGACTCACAGGGCTTTGTGCTCGATCATTGGAGACCCGGACCTTCGGAATTGA
Protein
MCVAFNKQSPRLYASGMTNWMRYQILLLHNRHRDNIAMGLEAGQPPATNMRKMYWDYELEEMAELWARQCQKRHDECRNTIRFNALQNMDVRPVVPKVTEAQLLADAVGAWFSGSRVLQSEHVWSFRHGNCSAPGGCNSHWYSTAAWASTWRLGCSQALCTATKKSCSRFRRRATPSEDLTNTSTRVTGENILRRTRYMTTATFPKFKDFATTVRVKTKESPLHRGILEMTNVRKANDEPKIVAFTFCNYGPAGNIDGYPIYEAGLSCSNCPQGTRCGDRTHRALCSIIGDPDLRN
Summary
Similarity
Belongs to the CRISP family.
Uniprot
EMBL
BABH01039778
KZ149977
PZC75907.1
NWSH01001381
PCG71464.1
KQ461196
+ More
KPJ06406.1 KQ459605 KPI91890.1 JTDY01002281 KOB71745.1 ODYU01002248 SOQ39479.1 AGBW02013269 OWR43743.1 GDIQ01104011 JAL47715.1 GDIP01019685 JAM84030.1 GDIQ01020988 JAN73749.1 LRGB01001581 KZS11094.1 GDIQ01062661 JAN32076.1 GDIQ01112498 JAL39228.1 GDIP01080043 JAM23672.1 AJVK01015784 GDIQ01134204 JAL17522.1 GFDF01005219 JAV08865.1
KPJ06406.1 KQ459605 KPI91890.1 JTDY01002281 KOB71745.1 ODYU01002248 SOQ39479.1 AGBW02013269 OWR43743.1 GDIQ01104011 JAL47715.1 GDIP01019685 JAM84030.1 GDIQ01020988 JAN73749.1 LRGB01001581 KZS11094.1 GDIQ01062661 JAN32076.1 GDIQ01112498 JAL39228.1 GDIP01080043 JAM23672.1 AJVK01015784 GDIQ01134204 JAL17522.1 GFDF01005219 JAV08865.1
Proteomes
PRIDE
Pfam
PF00188 CAP
Interpro
SUPFAM
SSF55797
SSF55797
Gene 3D
ProteinModelPortal
PDB
1QNX
E-value=6.24915e-12,
Score=169
Ontologies
GO
PANTHER
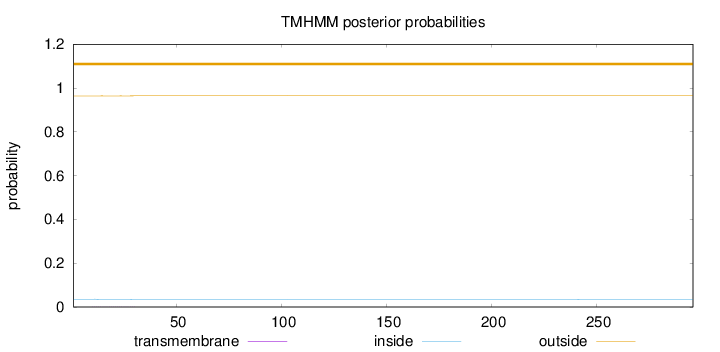
Topology
Length:
296
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.02037
Exp number, first 60 AAs:
0.01764
Total prob of N-in:
0.03592
outside
1 - 296
Population Genetic Test Statistics
Pi
13.214166
Theta
12.278892
Tajima's D
0.144105
CLR
0
CSRT
0.423028848557572
Interpretation
Uncertain
