Gene
KWMTBOMO00465
Annotation
PREDICTED:_TBC1_domain_family_member_13_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.73
Sequence
CDS
ATGAGTCTCCATAAAGCTAGGATAAAAGCCTTTGAAGAAGTACTTGATCAAGATGTTATTGATATGCAGCAGCTCCAGAAGCTAGCCTTCAATGGAATACCTGATGATAAAGGACTGAGATCATTAGTTTGGAAAATCCTACTGCATTATTTACCCCAAGAAAAGAACGCTAGAGAAACAACGCTAATTAAGAAACGTCAGCTTTATAAACAGTTTATAGAAGAAATAATTGTATCGCCCGGAGGGCCAACTGACCATCCGTTAAACATGAGCCCCGACAGCTCTTGGAGTACTTACTTCAAAGATAACGAGGTACTGTTGCAAATCGACAAGGACGTGCGAAGACTCTGCCCCGATATATCATTCTTTCAATCTGCTACTGAATTTCCTTGTCCTGAGGTCGTAAACAGCAATGGCATAAAACGATTGCACAAACGTGTCGAGCAATCCGTCCTCAAATACGCCACATTGGAGAGAAGGGGTCTCGGTGTTGCCAAGCTTAGCAACGAAATCAAACGCGCCGAGAACATCACAAGCGGCGATTACGCCCCACTGAACGAAGGCTGCGAGGCTCACTGGGAGGTGGTGGAACGTATGCTGTTTCTGTATGCGAAGCTGAACCCGGGACAGGGCTACGTTCAAGGAATGAACGAAATAATAGGACCCATCTATCATACCTTCGCTATTGACGCCGATAAGGAGTTTCGACAGCACGCCGAAGCCGACTGCTTCTTCTGCTTCACGAACCTAATGGCCGAGATACGTGACTTCTTCATACGTACCCTCGACGAGGCGGAAAGCGGAATCAACTACATGATGTGCAAACTCAACGAATTTCTGAAGCGCCACGACCCGGACGTTTGGACTTTATTGGAGAAGCAGGAACTGCGCCCTCAGTACTACAGCTTCAGATGGCTGACGCTCTTACTGTCCCAAGAGTTCTCTCTACCCGACGTCGAAAGAATTTGGGACACCCTATTCGCTGATTCTCAACGTTTCGATTTCCTCATAAACATATGCTGCGCGATGATTTTGTTGGTTCGGGATAATTTACTAGGCGGCGATTTCGCGTCGAATGTAAAATTGCTACAAAACTTTCCGCCTATGGACGTGTCGTTGATACTAAATAAAGCAGCTACACTCGGCAACACGTAA
Protein
MSLHKARIKAFEEVLDQDVIDMQQLQKLAFNGIPDDKGLRSLVWKILLHYLPQEKNARETTLIKKRQLYKQFIEEIIVSPGGPTDHPLNMSPDSSWSTYFKDNEVLLQIDKDVRRLCPDISFFQSATEFPCPEVVNSNGIKRLHKRVEQSVLKYATLERRGLGVAKLSNEIKRAENITSGDYAPLNEGCEAHWEVVERMLFLYAKLNPGQGYVQGMNEIIGPIYHTFAIDADKEFRQHAEADCFFCFTNLMAEIRDFFIRTLDEAESGINYMMCKLNEFLKRHDPDVWTLLEKQELRPQYYSFRWLTLLLSQEFSLPDVERIWDTLFADSQRFDFLINICCAMILLVRDNLLGGDFASNVKLLQNFPPMDVSLILNKAATLGNT
Summary
Uniprot
A0A212EQL2
A0A194QN51
A0A194PEV1
A0A2H1VF63
A0A2W1BQN4
H9JUW2
+ More
A0A1E1WR41 A0A2J7QK97 E9J167 D6WI52 A0A088AC46 A0A026W3G7 A0A1Y1NBB1 E2A9E6 A0A158N8Y1 A0A0L7RJZ0 A0A151ISD7 A0A1B6DHC2 A0A151WNP5 N6UI94 A0A151HZN5 F4W9H0 A0A195C982 A0A224XR45 A0A1B6LT36 A0A1W4WGH6 A0A0P4VSA1 A0A232F8M1 K7J4B9 E0V9S3 A0A1B6HGD4 E2C3W1 Q0IG78 T1I605 A0A182G8R9 A0A195F5Q5 A0A0C9RXS4 A0A182GPG3 A0A2A3EH27 A0A1Q3G1X5 A0A1Q3G205 A0A1S4F3K3 A0A0A9YDC1 A0A336M0R4 A0A1A9X358 A0A1I8PJY2 A0A0L0CKV4 A0A1B0GLY5 A0A0M9A702 B4PFZ6 A0A067QW59 A0A0R1DV69 A0A1I8MR39 B3NBQ7 Q9VSM8 A0A1W4US91 A0A3B0JVK6 A0A182WKW7 Q95TT8 A0A182SC41 A0A0J9RRI7 A0A0Q5UI42 A0A182RS09 A0A0J9RRY4 A0A1W4UEU7 A0A182JHM1 A0A0J9RRM4 A0A182UBU9 Q7Q6P6 A0A182HWK0 A0A1B0EZ32 B3M682 A0A182XMP3 A0A182YPJ6 B4N3P4 A0A182MEF8 A0A1L8DVU5 A0A182L329 A0A1W4WGE9 B4HK05 A0A1A9V2W8 A0A310S9N5 A0A182Q402 W8C6T1 U5EW07 A0A182NH51 A0A1A9XKA8 A0A182K386 B4H9I6 A0A182URC0 A0A1B0B471 A0A034W9S6 W5JGN1 A0A0K8V668 A0A1A9ZFW4 B5DRN7 B4LEL4 E9FUD3 B4KZ63 D3TL41
A0A1E1WR41 A0A2J7QK97 E9J167 D6WI52 A0A088AC46 A0A026W3G7 A0A1Y1NBB1 E2A9E6 A0A158N8Y1 A0A0L7RJZ0 A0A151ISD7 A0A1B6DHC2 A0A151WNP5 N6UI94 A0A151HZN5 F4W9H0 A0A195C982 A0A224XR45 A0A1B6LT36 A0A1W4WGH6 A0A0P4VSA1 A0A232F8M1 K7J4B9 E0V9S3 A0A1B6HGD4 E2C3W1 Q0IG78 T1I605 A0A182G8R9 A0A195F5Q5 A0A0C9RXS4 A0A182GPG3 A0A2A3EH27 A0A1Q3G1X5 A0A1Q3G205 A0A1S4F3K3 A0A0A9YDC1 A0A336M0R4 A0A1A9X358 A0A1I8PJY2 A0A0L0CKV4 A0A1B0GLY5 A0A0M9A702 B4PFZ6 A0A067QW59 A0A0R1DV69 A0A1I8MR39 B3NBQ7 Q9VSM8 A0A1W4US91 A0A3B0JVK6 A0A182WKW7 Q95TT8 A0A182SC41 A0A0J9RRI7 A0A0Q5UI42 A0A182RS09 A0A0J9RRY4 A0A1W4UEU7 A0A182JHM1 A0A0J9RRM4 A0A182UBU9 Q7Q6P6 A0A182HWK0 A0A1B0EZ32 B3M682 A0A182XMP3 A0A182YPJ6 B4N3P4 A0A182MEF8 A0A1L8DVU5 A0A182L329 A0A1W4WGE9 B4HK05 A0A1A9V2W8 A0A310S9N5 A0A182Q402 W8C6T1 U5EW07 A0A182NH51 A0A1A9XKA8 A0A182K386 B4H9I6 A0A182URC0 A0A1B0B471 A0A034W9S6 W5JGN1 A0A0K8V668 A0A1A9ZFW4 B5DRN7 B4LEL4 E9FUD3 B4KZ63 D3TL41
Pubmed
22118469
26354079
28756777
19121390
21282665
18362917
+ More
19820115 24508170 30249741 28004739 20798317 21347285 23537049 21719571 27129103 28648823 20075255 20566863 17510324 26483478 25401762 26823975 26108605 17994087 17550304 24845553 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 12364791 14747013 17210077 25244985 20966253 24495485 25348373 20920257 23761445 15632085 21292972 20353571
19820115 24508170 30249741 28004739 20798317 21347285 23537049 21719571 27129103 28648823 20075255 20566863 17510324 26483478 25401762 26823975 26108605 17994087 17550304 24845553 25315136 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 12364791 14747013 17210077 25244985 20966253 24495485 25348373 20920257 23761445 15632085 21292972 20353571
EMBL
AGBW02013269
OWR43744.1
KQ461196
KPJ06405.1
KQ459605
KPI91891.1
+ More
ODYU01002248 SOQ39478.1 KZ149977 PZC75905.1 BABH01039780 GDQN01001559 JAT89495.1 NEVH01013275 PNF29006.1 GL767586 EFZ13409.1 KQ971334 EFA01059.1 KK107453 QOIP01000004 EZA50578.1 RLU23589.1 GEZM01010199 JAV94150.1 GL437804 EFN69943.1 ADTU01008907 KQ414574 KOC71267.1 KQ981075 KYN09782.1 GEDC01012224 JAS25074.1 KQ982907 KYQ49411.1 APGK01019133 KB740092 KB632429 ENN81455.1 ERL95437.1 KQ976714 KYM76943.1 GL888030 EGI69145.1 KQ978068 KYM97382.1 GFTR01005945 JAW10481.1 GEBQ01013129 JAT26848.1 GDKW01001790 JAI54805.1 NNAY01000718 OXU26818.1 AAZX01006389 DS234996 EEB10129.1 GECU01033984 JAS73722.1 GL452364 EFN77347.1 CH477261 EAT45679.1 ACPB03000757 JXUM01009565 KQ560286 KXJ83268.1 KQ981810 KYN35404.1 GBYB01014025 GBYB01014027 JAG83792.1 JAG83794.1 JXUM01078413 KQ563065 KXJ74544.1 KZ288258 PBC30519.1 GFDL01001230 JAV33815.1 GFDL01001213 JAV33832.1 GBHO01013998 GDHC01004973 JAG29606.1 JAQ13656.1 UFQT01000224 SSX22017.1 JRES01000247 KNC32998.1 AJVK01020104 AJVK01020105 AJVK01020106 KQ435735 KOX77149.1 CM000159 EDW93140.1 KK853300 KDR08764.1 KRK00969.1 CH954178 EDV50653.1 AE014296 BT044356 AAF50388.1 ACH92421.1 OUUW01000002 SPP77396.1 AY058542 AAL13771.1 CM002912 KMY98431.1 KQS43426.1 KMY98432.1 KMY98433.1 AAAB01008960 EAA11714.4 APCN01001496 AJWK01021800 CH902618 EDV39702.1 CH964095 EDW79249.1 AXCM01013222 GFDF01003534 JAV10550.1 CH480815 EDW40741.1 KQ766819 OAD53558.1 AXCN02000346 GAMC01000947 JAC05609.1 GANO01001636 JAB58235.1 CH479229 EDW36462.1 JXJN01008181 GAKP01008419 JAC50533.1 ADMH02001558 ETN62070.1 GDHF01017885 JAI34429.1 CH379070 EDY74272.2 CH940647 EDW69099.1 GL732525 EFX88939.1 CH933809 EDW17860.1 EZ422139 ADD18336.1
ODYU01002248 SOQ39478.1 KZ149977 PZC75905.1 BABH01039780 GDQN01001559 JAT89495.1 NEVH01013275 PNF29006.1 GL767586 EFZ13409.1 KQ971334 EFA01059.1 KK107453 QOIP01000004 EZA50578.1 RLU23589.1 GEZM01010199 JAV94150.1 GL437804 EFN69943.1 ADTU01008907 KQ414574 KOC71267.1 KQ981075 KYN09782.1 GEDC01012224 JAS25074.1 KQ982907 KYQ49411.1 APGK01019133 KB740092 KB632429 ENN81455.1 ERL95437.1 KQ976714 KYM76943.1 GL888030 EGI69145.1 KQ978068 KYM97382.1 GFTR01005945 JAW10481.1 GEBQ01013129 JAT26848.1 GDKW01001790 JAI54805.1 NNAY01000718 OXU26818.1 AAZX01006389 DS234996 EEB10129.1 GECU01033984 JAS73722.1 GL452364 EFN77347.1 CH477261 EAT45679.1 ACPB03000757 JXUM01009565 KQ560286 KXJ83268.1 KQ981810 KYN35404.1 GBYB01014025 GBYB01014027 JAG83792.1 JAG83794.1 JXUM01078413 KQ563065 KXJ74544.1 KZ288258 PBC30519.1 GFDL01001230 JAV33815.1 GFDL01001213 JAV33832.1 GBHO01013998 GDHC01004973 JAG29606.1 JAQ13656.1 UFQT01000224 SSX22017.1 JRES01000247 KNC32998.1 AJVK01020104 AJVK01020105 AJVK01020106 KQ435735 KOX77149.1 CM000159 EDW93140.1 KK853300 KDR08764.1 KRK00969.1 CH954178 EDV50653.1 AE014296 BT044356 AAF50388.1 ACH92421.1 OUUW01000002 SPP77396.1 AY058542 AAL13771.1 CM002912 KMY98431.1 KQS43426.1 KMY98432.1 KMY98433.1 AAAB01008960 EAA11714.4 APCN01001496 AJWK01021800 CH902618 EDV39702.1 CH964095 EDW79249.1 AXCM01013222 GFDF01003534 JAV10550.1 CH480815 EDW40741.1 KQ766819 OAD53558.1 AXCN02000346 GAMC01000947 JAC05609.1 GANO01001636 JAB58235.1 CH479229 EDW36462.1 JXJN01008181 GAKP01008419 JAC50533.1 ADMH02001558 ETN62070.1 GDHF01017885 JAI34429.1 CH379070 EDY74272.2 CH940647 EDW69099.1 GL732525 EFX88939.1 CH933809 EDW17860.1 EZ422139 ADD18336.1
Proteomes
UP000007151
UP000053240
UP000053268
UP000005204
UP000235965
UP000007266
+ More
UP000005203 UP000053097 UP000279307 UP000000311 UP000005205 UP000053825 UP000078492 UP000075809 UP000019118 UP000030742 UP000078540 UP000007755 UP000078542 UP000192223 UP000215335 UP000002358 UP000009046 UP000008237 UP000008820 UP000015103 UP000069940 UP000249989 UP000078541 UP000242457 UP000091820 UP000095300 UP000037069 UP000092462 UP000053105 UP000002282 UP000027135 UP000095301 UP000008711 UP000000803 UP000192221 UP000268350 UP000075920 UP000075901 UP000075900 UP000075880 UP000075902 UP000007062 UP000075840 UP000092461 UP000007801 UP000076407 UP000076408 UP000007798 UP000075883 UP000075882 UP000001292 UP000078200 UP000075886 UP000075884 UP000092443 UP000075881 UP000008744 UP000075903 UP000092460 UP000000673 UP000092445 UP000001819 UP000008792 UP000000305 UP000009192
UP000005203 UP000053097 UP000279307 UP000000311 UP000005205 UP000053825 UP000078492 UP000075809 UP000019118 UP000030742 UP000078540 UP000007755 UP000078542 UP000192223 UP000215335 UP000002358 UP000009046 UP000008237 UP000008820 UP000015103 UP000069940 UP000249989 UP000078541 UP000242457 UP000091820 UP000095300 UP000037069 UP000092462 UP000053105 UP000002282 UP000027135 UP000095301 UP000008711 UP000000803 UP000192221 UP000268350 UP000075920 UP000075901 UP000075900 UP000075880 UP000075902 UP000007062 UP000075840 UP000092461 UP000007801 UP000076407 UP000076408 UP000007798 UP000075883 UP000075882 UP000001292 UP000078200 UP000075886 UP000075884 UP000092443 UP000075881 UP000008744 UP000075903 UP000092460 UP000000673 UP000092445 UP000001819 UP000008792 UP000000305 UP000009192
Pfam
PF00566 RabGAP-TBC
SUPFAM
SSF47923
SSF47923
ProteinModelPortal
A0A212EQL2
A0A194QN51
A0A194PEV1
A0A2H1VF63
A0A2W1BQN4
H9JUW2
+ More
A0A1E1WR41 A0A2J7QK97 E9J167 D6WI52 A0A088AC46 A0A026W3G7 A0A1Y1NBB1 E2A9E6 A0A158N8Y1 A0A0L7RJZ0 A0A151ISD7 A0A1B6DHC2 A0A151WNP5 N6UI94 A0A151HZN5 F4W9H0 A0A195C982 A0A224XR45 A0A1B6LT36 A0A1W4WGH6 A0A0P4VSA1 A0A232F8M1 K7J4B9 E0V9S3 A0A1B6HGD4 E2C3W1 Q0IG78 T1I605 A0A182G8R9 A0A195F5Q5 A0A0C9RXS4 A0A182GPG3 A0A2A3EH27 A0A1Q3G1X5 A0A1Q3G205 A0A1S4F3K3 A0A0A9YDC1 A0A336M0R4 A0A1A9X358 A0A1I8PJY2 A0A0L0CKV4 A0A1B0GLY5 A0A0M9A702 B4PFZ6 A0A067QW59 A0A0R1DV69 A0A1I8MR39 B3NBQ7 Q9VSM8 A0A1W4US91 A0A3B0JVK6 A0A182WKW7 Q95TT8 A0A182SC41 A0A0J9RRI7 A0A0Q5UI42 A0A182RS09 A0A0J9RRY4 A0A1W4UEU7 A0A182JHM1 A0A0J9RRM4 A0A182UBU9 Q7Q6P6 A0A182HWK0 A0A1B0EZ32 B3M682 A0A182XMP3 A0A182YPJ6 B4N3P4 A0A182MEF8 A0A1L8DVU5 A0A182L329 A0A1W4WGE9 B4HK05 A0A1A9V2W8 A0A310S9N5 A0A182Q402 W8C6T1 U5EW07 A0A182NH51 A0A1A9XKA8 A0A182K386 B4H9I6 A0A182URC0 A0A1B0B471 A0A034W9S6 W5JGN1 A0A0K8V668 A0A1A9ZFW4 B5DRN7 B4LEL4 E9FUD3 B4KZ63 D3TL41
A0A1E1WR41 A0A2J7QK97 E9J167 D6WI52 A0A088AC46 A0A026W3G7 A0A1Y1NBB1 E2A9E6 A0A158N8Y1 A0A0L7RJZ0 A0A151ISD7 A0A1B6DHC2 A0A151WNP5 N6UI94 A0A151HZN5 F4W9H0 A0A195C982 A0A224XR45 A0A1B6LT36 A0A1W4WGH6 A0A0P4VSA1 A0A232F8M1 K7J4B9 E0V9S3 A0A1B6HGD4 E2C3W1 Q0IG78 T1I605 A0A182G8R9 A0A195F5Q5 A0A0C9RXS4 A0A182GPG3 A0A2A3EH27 A0A1Q3G1X5 A0A1Q3G205 A0A1S4F3K3 A0A0A9YDC1 A0A336M0R4 A0A1A9X358 A0A1I8PJY2 A0A0L0CKV4 A0A1B0GLY5 A0A0M9A702 B4PFZ6 A0A067QW59 A0A0R1DV69 A0A1I8MR39 B3NBQ7 Q9VSM8 A0A1W4US91 A0A3B0JVK6 A0A182WKW7 Q95TT8 A0A182SC41 A0A0J9RRI7 A0A0Q5UI42 A0A182RS09 A0A0J9RRY4 A0A1W4UEU7 A0A182JHM1 A0A0J9RRM4 A0A182UBU9 Q7Q6P6 A0A182HWK0 A0A1B0EZ32 B3M682 A0A182XMP3 A0A182YPJ6 B4N3P4 A0A182MEF8 A0A1L8DVU5 A0A182L329 A0A1W4WGE9 B4HK05 A0A1A9V2W8 A0A310S9N5 A0A182Q402 W8C6T1 U5EW07 A0A182NH51 A0A1A9XKA8 A0A182K386 B4H9I6 A0A182URC0 A0A1B0B471 A0A034W9S6 W5JGN1 A0A0K8V668 A0A1A9ZFW4 B5DRN7 B4LEL4 E9FUD3 B4KZ63 D3TL41
PDB
2QFZ
E-value=5.47968e-18,
Score=223
Ontologies
GO
Topology
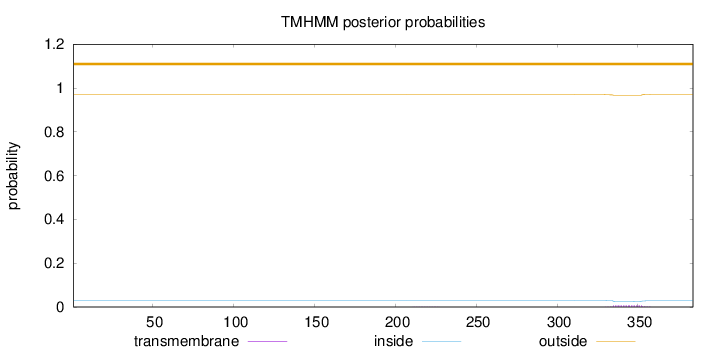
Length:
384
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.15612
Exp number, first 60 AAs:
0
Total prob of N-in:
0.03009
outside
1 - 384
Population Genetic Test Statistics
Pi
16.934868
Theta
17.401807
Tajima's D
-0.993638
CLR
0.043277
CSRT
0.137593120343983
Interpretation
Uncertain
