Gene
KWMTBOMO00462 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013328
Annotation
tudor_staphylococcus/micrococcal_nuclease_[Bombyx_mori]
Full name
Staphylococcal nuclease domain-containing protein
+ More
Staphylococcal nuclease domain-containing protein 1
Staphylococcal nuclease domain-containing protein 1
Alternative Name
Tudor staphylococcal nuclease
Location in the cell
Cytoplasmic Reliability : 1.87 Mitochondrial Reliability : 1.382
Sequence
CDS
ATGAGTGCTCCAGCACCGGCACCTGCTTACAAAATAGGAATAGTGAAGCAGGTACTATCGGGAGATACGGTGGTAATAAGAAAACAGCCTCAGGGCGGGCCGCCTCCAGAGAAAGTGATAGCCCTTTCGGGTATCACCGCACCTAAACTCGCTCGCCAAAGGACCGCCAACAATGACACAGAAACGAAGGATGAACCATTCGCGTGGGAAGCCCGTGAATTCCTAAGGAAGAAGCTAGTCGGCAAGGAGGTTATCTTCACTGCTGAGAAACCGGCGAATTCAGCGAATAGAGAATATGGCGTCGTATGGGCTGGCAAAAATCCTTCCAAAGATGAAAATGTTACTGAAGCCTTGCTGGCTGAAGGTCTCGTTAAAGTTCGAGAAGGTGCTAGAAATATACCTCAGCTCAAAAAGCTGGTAGAGATCGAAGAGACAGCCAAATCGCAAGGCAAAGGAATTTGGAGTACTGATGCTGATAAACATGTACGAGACATCAAGTGGTCTATCGAAAATCTGAAAGCTTTCGTCAACAAATACAATGGTAAGCCTGTAAAGGCTATCATTGAATATGTGCGTGATGGTTCTACAGTACGTCTTTGTTTGTTACCTGAATACACTCCCATTACACTGATGCTATCTGGAATTCGATGTCCCGCTGTAAGACAAGACGGTGAATCTGAGCCTTACGCAGAAGAAGCACGCTTCTTCCTGGAGTCCAGATTGCTGCAGAAAGACGTCGAAGTAGTTCTGGAGTCCGTTAATAACAACAATATCGTGGGCACGATACTGCACCCGCAGGGTAACATCGCTGAGGCCCTACTAAGACAAGGGTTCGCGAAATGTGTCATGAAATCAGGTGCATCTACATTGAAAGCCGCCGAAAGTGCAGCTAAGGAAGCAAAACTGCGCATATGGATAAATTATGTAAGCAATGCACCTATAATCCCGGCGAAAGACAAGGAATTCACGGCCACCGTGTTGGAAGTAGTCAACGGAGACGCGCTCGTTGTTAAAACACATAGCAATGTACAGAAAAAAATATTCTTAGCTAGCGTCAGGCCTCCTCGTGAAAAGAGCAATGCTGACGACGACAATAAACCTATACCGAGGCCAAAAGGATTTAGACCTTTGTATGATATTCCATGGATGTACGAGGCACGTGAATTTCTTAGAAAAAAACTGGTTGGGAAAAAAGTTAATGTGACAGTAGATTACATTCAGCCAGCAAAAGATAATTTCCCAGAAAAGACATGTTGTACTGTCGTAAGCGGTGGCACGAACATAGCTGAAGCTCTGGTGACTAAAGGGTTGGCAACAGTGAAGTACAGAAACGATAATGACCAAAGAAGCTCACAGTATGATAAATTATTGGAGGCTGAATTGAAAGCAGTTAAGGCCGCAGTTGGTATTCATAACAAGAAAGAAGTGCCCACCCACCGTGTGCAAGACACCAGTGGAGATCCAACGAAAGCAAAAAAATTCTTCCCATTCCTCAAACGTGCCCAAAAAACCGAGGCTACTGTGGAATTCGTTGCTAGCGGATCGCGTATGCGTTTATATATTCCTAAAGAATCCGTCTTAGTAACCTTTTTGCTTGCCGGCATTAACTGTCCCCGAGGTGCGAGACCTGCAATTGGCGGAGGACCCAAACAAAATGCAGAGCCTTTCGGAGAAGAAGCTTTGCAGTTCACGAAAGAAAAGTGCTTACAGCACGACGTCTTAGTCAGCATAGAAGAGATTGACAAAGCAGGAAACTTCATCGGTTGGCTCTGGGTTGACAACGAGAATTTGTCCGTTTCGTTAGTCGAACATGGCCTTGCCTCGGCGCATCACACGGCGGAAACGTCTGAGTTTGCACGCGCCATAAAAACCGCTGAAGAGAATGCAATTAAAAAGCGTATCGGCGTATGGAAAGACTATGTCGAAGAAGAAAAAGAAGTCGAAAAGGAGAGAAATGCAACCGTACAAGATCGTACTCTTAAATACGACCGCGTGGTCATCACTGAAGTCACTCCTGAAGGTCATTTCTACGCACAAAATGTCGATTTAGGTGTGAAACTAGAAAGTCTTATGGAAACTATACACACTGAGTTTAGAAACAGTCACCCATTGCCTGGATCTTACGCTCCAAGGAAGGGAGCCATCTGTGCTGCTCGTTTTACAGCCGATGACCAATGGTACAGGGCAAAGATTGAAAAAATAACAGACAATAGGCAGGTCCAAGTTGTATACATCGACTATGGAAATCGAGAGACGTTAGACATAACCAGATTGGCAGCACTCCCGGTGGGTACTGAGCACGATTCGCCTTTTGCGACAGAGTATGTTCTCTGCTGTGTGAAATTCCCCTCGGACCCTGATGACCGAGCAGAAGCTGTAACCAGTTTCTATAATGATGTCGTTGGCAAGAGGCTGCTCTTGAATGTAGAAATCCGTGGATCACCAGCTGCTGTTACCCTTGTTGATCCTAACACAAACATTGATCTGGGCAAGAACCTTATCAAAGATGGGCTCGTGTTAGTGGAGCAGGTTCGTGACAGCCGACTGGCAACATTGATGGCCGAGTATCGCGCCGCTCAGGAGCACGCCAAGAGCTCGCGACTCAACTTGTGGCGGCACGGTGACATCACCGAGGACGATGCGGTAGAATTCGGGGTGCGTCGCTAG
Protein
MSAPAPAPAYKIGIVKQVLSGDTVVIRKQPQGGPPPEKVIALSGITAPKLARQRTANNDTETKDEPFAWEAREFLRKKLVGKEVIFTAEKPANSANREYGVVWAGKNPSKDENVTEALLAEGLVKVREGARNIPQLKKLVEIEETAKSQGKGIWSTDADKHVRDIKWSIENLKAFVNKYNGKPVKAIIEYVRDGSTVRLCLLPEYTPITLMLSGIRCPAVRQDGESEPYAEEARFFLESRLLQKDVEVVLESVNNNNIVGTILHPQGNIAEALLRQGFAKCVMKSGASTLKAAESAAKEAKLRIWINYVSNAPIIPAKDKEFTATVLEVVNGDALVVKTHSNVQKKIFLASVRPPREKSNADDDNKPIPRPKGFRPLYDIPWMYEAREFLRKKLVGKKVNVTVDYIQPAKDNFPEKTCCTVVSGGTNIAEALVTKGLATVKYRNDNDQRSSQYDKLLEAELKAVKAAVGIHNKKEVPTHRVQDTSGDPTKAKKFFPFLKRAQKTEATVEFVASGSRMRLYIPKESVLVTFLLAGINCPRGARPAIGGGPKQNAEPFGEEALQFTKEKCLQHDVLVSIEEIDKAGNFIGWLWVDNENLSVSLVEHGLASAHHTAETSEFARAIKTAEENAIKKRIGVWKDYVEEEKEVEKERNATVQDRTLKYDRVVITEVTPEGHFYAQNVDLGVKLESLMETIHTEFRNSHPLPGSYAPRKGAICAARFTADDQWYRAKIEKITDNRQVQVVYIDYGNRETLDITRLAALPVGTEHDSPFATEYVLCCVKFPSDPDDRAEAVTSFYNDVVGKRLLLNVEIRGSPAAVTLVDPNTNIDLGKNLIKDGLVLVEQVRDSRLATLMAEYRAAQEHAKSSRLNLWRHGDITEDDAVEFGVRR
Summary
Description
Endonuclease that mediates miRNA decay of both protein-free and AGO2-loaded miRNAs.
Endonuclease which shows activity towards both DNA and RNA substrates (PubMed:14508492, PubMed:26808625). Has a role in translation regulation throught its association with the with the RNA-induced silencing complex (RISC) (PubMed:14508492, PubMed:26808625). Plays a role in spermatogenesis probably by negatively regulating piwi expression in the germline (PubMed:26808625). Together with piwi, might be involved in transposon repression in the germline (PubMed:26808625).
Endonuclease which shows activity towards both DNA and RNA substrates (PubMed:14508492, PubMed:26808625). Has a role in translation regulation throught its association with the with the RNA-induced silencing complex (RISC) (PubMed:14508492, PubMed:26808625). Plays a role in spermatogenesis probably by negatively regulating piwi expression in the germline (PubMed:26808625). Together with piwi, might be involved in transposon repression in the germline (PubMed:26808625).
Catalytic Activity
Endonucleolytic cleavage to nucleoside 3'-phosphates and 3'-phosphooligonucleotide end-products.
Subunit
Associates with the RNA-induced silencing complex (RISC) (PubMed:14508492). Interacts with the RISC components AGO2, Fmr1 and vig (PubMed:14508492). Interacts with piwi (PubMed:26808625).
Keywords
3D-structure
Complete proteome
Cytoplasm
Endonuclease
Hydrolase
Nuclease
Nucleus
Reference proteome
Repeat
RNA-binding
RNA-mediated gene silencing
Feature
chain Staphylococcal nuclease domain-containing protein
Uniprot
D9N4J4
H9JUW5
A0A2W1BLH6
A0A212EQK0
S4PY67
A0A2H1W6B7
+ More
A0A2A4J4G7 A0A194QN47 A0A194PF40 A0A2J7RK75 A0A2J7RK72 A0A067RA05 A0A1L7NZM8 A0A1Y1NAS3 A0A0P5RPB4 A0A0P6ECC7 A0A0P6D5Y7 A0A0P5JM29 D6WH29 A0A0P5M5M6 A0A0P6DAC5 A0A0T6AYP2 A0A0P5NMF8 A0A0P5MWB4 A0A0P6DA35 A0A0P6F8J1 A0A0P6IWG6 A0A0P5VNB2 A0A0P5BRP3 A0A0P5UMH7 A0A0P5IFM4 A0A0P5BRR9 A0A0P5Y6Z4 A0A0N8C6G6 A0A0N8B3V9 Q17PM3 A0A0P4Z1P6 A0A023EY38 A0A126Q9D5 A0A182H564 E9GT82 A0A1W4XCF9 K1R512 A0A0N8DHY8 A0A1B6EL67 A0A0P5XQE1 U5EY13 A0A0P5HPX3 A0A1S3IS83 A0A0P5G4Z2 V3ZN51 A0A0P5KVE5 A0A0N8D1M8 A0A0P5SXH0 A0A1B6ECM6 B0WIK3 A0A1Q3F1Q3 A0A0P5RCA0 A0A1Q3F1R2 A0A210Q8D8 A0A0P5IES6 A0A0P5RAE9 A0A2C9JNE9 A0A1Z5LGF5 A0A0P4ZBL9 A0A0P5AQQ8 A0A2R5LE81 A0A0P6G5Q7 A0A0P6EE22 A0A1L8DGR0 G1ARD5 A0A0P5K1F1 A0A0P5VBJ5 A0A0P5VEH1 A0A0B6ZVN1 A0A0N8CGJ0 A0A1J1IXN9 A0A2I9LPS2 A0A087ULG5 A0A0P5QNC1 A0A0P4XWZ0 A0A0P5IIZ5 A7RXU0 A0A1I8N1Y7 N1PBE4 A0A0P5R6M2 A0A0P4ZZC7 A0A0A1WIC5 A0A1W4VNR6 T1PB73 A0A0P4Z5X6 B4KXY0 B4MGQ8 B4QEY1 A0A0L8GAX7 A0A0M3QVT8 N6TSV1 Q9W0S7 A0A1I8PTZ6 B3NEM9
A0A2A4J4G7 A0A194QN47 A0A194PF40 A0A2J7RK75 A0A2J7RK72 A0A067RA05 A0A1L7NZM8 A0A1Y1NAS3 A0A0P5RPB4 A0A0P6ECC7 A0A0P6D5Y7 A0A0P5JM29 D6WH29 A0A0P5M5M6 A0A0P6DAC5 A0A0T6AYP2 A0A0P5NMF8 A0A0P5MWB4 A0A0P6DA35 A0A0P6F8J1 A0A0P6IWG6 A0A0P5VNB2 A0A0P5BRP3 A0A0P5UMH7 A0A0P5IFM4 A0A0P5BRR9 A0A0P5Y6Z4 A0A0N8C6G6 A0A0N8B3V9 Q17PM3 A0A0P4Z1P6 A0A023EY38 A0A126Q9D5 A0A182H564 E9GT82 A0A1W4XCF9 K1R512 A0A0N8DHY8 A0A1B6EL67 A0A0P5XQE1 U5EY13 A0A0P5HPX3 A0A1S3IS83 A0A0P5G4Z2 V3ZN51 A0A0P5KVE5 A0A0N8D1M8 A0A0P5SXH0 A0A1B6ECM6 B0WIK3 A0A1Q3F1Q3 A0A0P5RCA0 A0A1Q3F1R2 A0A210Q8D8 A0A0P5IES6 A0A0P5RAE9 A0A2C9JNE9 A0A1Z5LGF5 A0A0P4ZBL9 A0A0P5AQQ8 A0A2R5LE81 A0A0P6G5Q7 A0A0P6EE22 A0A1L8DGR0 G1ARD5 A0A0P5K1F1 A0A0P5VBJ5 A0A0P5VEH1 A0A0B6ZVN1 A0A0N8CGJ0 A0A1J1IXN9 A0A2I9LPS2 A0A087ULG5 A0A0P5QNC1 A0A0P4XWZ0 A0A0P5IIZ5 A7RXU0 A0A1I8N1Y7 N1PBE4 A0A0P5R6M2 A0A0P4ZZC7 A0A0A1WIC5 A0A1W4VNR6 T1PB73 A0A0P4Z5X6 B4KXY0 B4MGQ8 B4QEY1 A0A0L8GAX7 A0A0M3QVT8 N6TSV1 Q9W0S7 A0A1I8PTZ6 B3NEM9
EC Number
3.1.31.1
Pubmed
19121390
28756777
22118469
23622113
26354079
24845553
+ More
28034629 28004739 18362917 19820115 17510324 24945155 26483478 21292972 22992520 23254933 28812685 15562597 28528879 21745576 29248469 17615350 25315136 25830018 17994087 18057021 22936249 23537049 10731132 12537572 12537569 14508492 26808625 19232356
28034629 28004739 18362917 19820115 17510324 24945155 26483478 21292972 22992520 23254933 28812685 15562597 28528879 21745576 29248469 17615350 25315136 25830018 17994087 18057021 22936249 23537049 10731132 12537572 12537569 14508492 26808625 19232356
EMBL
AB573814
BAJ14101.1
BABH01039784
KZ149977
PZC75899.1
AGBW02013269
+ More
OWR43749.1 GAIX01003818 JAA88742.1 ODYU01006633 SOQ48645.1 NWSH01003445 PCG66282.1 KQ461196 KPJ06400.1 KQ459605 KPI91897.1 NEVH01002981 PNF41237.1 PNF41236.1 KK852598 KDR20540.1 LC194150 BAW35372.1 GEZM01007667 JAV95011.1 GDIQ01097934 GDIQ01097933 JAL53793.1 GDIQ01189943 GDIQ01189942 GDIQ01065491 JAN29246.1 GDIQ01082634 GDIQ01082633 JAN12104.1 GDIQ01202950 JAK48775.1 KQ971321 EFA00120.1 GDIQ01160459 GDIQ01160458 JAK91267.1 GDIQ01216742 GDIQ01081079 JAN13658.1 LJIG01022488 KRT80307.1 GDIQ01140261 JAL11465.1 GDIQ01158668 JAK93057.1 GDIQ01081080 JAN13657.1 GDIQ01064049 GDIQ01064048 JAN30688.1 GDIQ01003523 JAN91214.1 GDIP01146872 GDIP01146871 GDIP01112182 GDIP01043182 LRGB01002451 JAL91532.1 KZS07676.1 GDIP01181026 JAJ42376.1 GDIP01110926 JAL92788.1 GDIQ01217840 GDIQ01025906 JAK33885.1 GDIP01185375 GDIQ01111869 JAJ38027.1 JAL39857.1 GDIP01200461 GDIP01062040 JAM41675.1 GDIQ01110131 JAL41595.1 GDIQ01215387 JAK36338.1 CH477190 EAT48729.1 GDIP01219015 JAJ04387.1 GAPW01000149 JAC13449.1 KT694370 AMK01822.1 JXUM01110873 JXUM01110874 JXUM01110875 KQ565451 KXJ70956.1 GL732563 EFX77404.1 JH818113 EKC40848.1 GDIP01031943 JAM71772.1 GECZ01031128 JAS38641.1 GDIP01068776 JAM34939.1 GANO01000655 JAB59216.1 GDIQ01226537 JAK25188.1 GDIQ01247185 JAK04540.1 KB203251 ESO85752.1 GDIQ01180052 JAK71673.1 GDIP01077623 JAM26092.1 GDIP01133928 JAL69786.1 GEDC01001617 JAS35681.1 DS231949 EDS28526.1 GFDL01013559 JAV21486.1 GDIQ01111871 JAL39855.1 GFDL01013550 JAV21495.1 NEDP02004628 OWF45007.1 GDIQ01215389 JAK36336.1 GDIQ01110130 JAL41596.1 GFJQ02000580 JAW06390.1 GDIP01219067 JAJ04335.1 GDIP01209815 JAJ13587.1 GGLE01003676 MBY07802.1 GDIQ01048909 JAN45828.1 GDIQ01064051 JAN30686.1 GFDF01008442 JAV05642.1 JF429696 AEK49107.1 GDIQ01196186 JAK55539.1 GDIP01117522 JAL86192.1 GDIP01116233 JAL87481.1 HACG01025526 HACG01025527 CEK72391.1 CEK72392.1 GDIP01133927 JAL69787.1 CVRI01000059 CRL03321.1 GFWZ01000395 MBW20385.1 KK120390 KFM78204.1 GDIQ01111872 JAL39854.1 GDIP01237824 JAI85577.1 GDIQ01215390 JAK36335.1 DS469551 EDO43681.1 AMQN01000055 KB291798 ELU18909.1 GDIQ01110128 JAL41598.1 GDIP01219068 JAJ04334.1 GBXI01015513 JAC98778.1 KA645996 AFP60625.1 GDIP01219066 JAJ04336.1 CH933809 EDW17652.1 CH940682 EDW57124.1 CM000362 CM002912 EDX07008.1 KMY96614.1 KQ422894 KOF74019.1 CP012525 ALC42969.1 APGK01055648 KB741269 ENN71421.1 AE014296 AY069361 BT003270 AAL39506.1 AAO25027.1 CH954178 EDV50024.1
OWR43749.1 GAIX01003818 JAA88742.1 ODYU01006633 SOQ48645.1 NWSH01003445 PCG66282.1 KQ461196 KPJ06400.1 KQ459605 KPI91897.1 NEVH01002981 PNF41237.1 PNF41236.1 KK852598 KDR20540.1 LC194150 BAW35372.1 GEZM01007667 JAV95011.1 GDIQ01097934 GDIQ01097933 JAL53793.1 GDIQ01189943 GDIQ01189942 GDIQ01065491 JAN29246.1 GDIQ01082634 GDIQ01082633 JAN12104.1 GDIQ01202950 JAK48775.1 KQ971321 EFA00120.1 GDIQ01160459 GDIQ01160458 JAK91267.1 GDIQ01216742 GDIQ01081079 JAN13658.1 LJIG01022488 KRT80307.1 GDIQ01140261 JAL11465.1 GDIQ01158668 JAK93057.1 GDIQ01081080 JAN13657.1 GDIQ01064049 GDIQ01064048 JAN30688.1 GDIQ01003523 JAN91214.1 GDIP01146872 GDIP01146871 GDIP01112182 GDIP01043182 LRGB01002451 JAL91532.1 KZS07676.1 GDIP01181026 JAJ42376.1 GDIP01110926 JAL92788.1 GDIQ01217840 GDIQ01025906 JAK33885.1 GDIP01185375 GDIQ01111869 JAJ38027.1 JAL39857.1 GDIP01200461 GDIP01062040 JAM41675.1 GDIQ01110131 JAL41595.1 GDIQ01215387 JAK36338.1 CH477190 EAT48729.1 GDIP01219015 JAJ04387.1 GAPW01000149 JAC13449.1 KT694370 AMK01822.1 JXUM01110873 JXUM01110874 JXUM01110875 KQ565451 KXJ70956.1 GL732563 EFX77404.1 JH818113 EKC40848.1 GDIP01031943 JAM71772.1 GECZ01031128 JAS38641.1 GDIP01068776 JAM34939.1 GANO01000655 JAB59216.1 GDIQ01226537 JAK25188.1 GDIQ01247185 JAK04540.1 KB203251 ESO85752.1 GDIQ01180052 JAK71673.1 GDIP01077623 JAM26092.1 GDIP01133928 JAL69786.1 GEDC01001617 JAS35681.1 DS231949 EDS28526.1 GFDL01013559 JAV21486.1 GDIQ01111871 JAL39855.1 GFDL01013550 JAV21495.1 NEDP02004628 OWF45007.1 GDIQ01215389 JAK36336.1 GDIQ01110130 JAL41596.1 GFJQ02000580 JAW06390.1 GDIP01219067 JAJ04335.1 GDIP01209815 JAJ13587.1 GGLE01003676 MBY07802.1 GDIQ01048909 JAN45828.1 GDIQ01064051 JAN30686.1 GFDF01008442 JAV05642.1 JF429696 AEK49107.1 GDIQ01196186 JAK55539.1 GDIP01117522 JAL86192.1 GDIP01116233 JAL87481.1 HACG01025526 HACG01025527 CEK72391.1 CEK72392.1 GDIP01133927 JAL69787.1 CVRI01000059 CRL03321.1 GFWZ01000395 MBW20385.1 KK120390 KFM78204.1 GDIQ01111872 JAL39854.1 GDIP01237824 JAI85577.1 GDIQ01215390 JAK36335.1 DS469551 EDO43681.1 AMQN01000055 KB291798 ELU18909.1 GDIQ01110128 JAL41598.1 GDIP01219068 JAJ04334.1 GBXI01015513 JAC98778.1 KA645996 AFP60625.1 GDIP01219066 JAJ04336.1 CH933809 EDW17652.1 CH940682 EDW57124.1 CM000362 CM002912 EDX07008.1 KMY96614.1 KQ422894 KOF74019.1 CP012525 ALC42969.1 APGK01055648 KB741269 ENN71421.1 AE014296 AY069361 BT003270 AAL39506.1 AAO25027.1 CH954178 EDV50024.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000053240
UP000053268
UP000235965
+ More
UP000027135 UP000007266 UP000076858 UP000008820 UP000069940 UP000249989 UP000000305 UP000192223 UP000005408 UP000085678 UP000030746 UP000002320 UP000242188 UP000076420 UP000183832 UP000054359 UP000001593 UP000095301 UP000014760 UP000192221 UP000009192 UP000008792 UP000000304 UP000053454 UP000092553 UP000019118 UP000000803 UP000095300 UP000008711
UP000027135 UP000007266 UP000076858 UP000008820 UP000069940 UP000249989 UP000000305 UP000192223 UP000005408 UP000085678 UP000030746 UP000002320 UP000242188 UP000076420 UP000183832 UP000054359 UP000001593 UP000095301 UP000014760 UP000192221 UP000009192 UP000008792 UP000000304 UP000053454 UP000092553 UP000019118 UP000000803 UP000095300 UP000008711
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
D9N4J4
H9JUW5
A0A2W1BLH6
A0A212EQK0
S4PY67
A0A2H1W6B7
+ More
A0A2A4J4G7 A0A194QN47 A0A194PF40 A0A2J7RK75 A0A2J7RK72 A0A067RA05 A0A1L7NZM8 A0A1Y1NAS3 A0A0P5RPB4 A0A0P6ECC7 A0A0P6D5Y7 A0A0P5JM29 D6WH29 A0A0P5M5M6 A0A0P6DAC5 A0A0T6AYP2 A0A0P5NMF8 A0A0P5MWB4 A0A0P6DA35 A0A0P6F8J1 A0A0P6IWG6 A0A0P5VNB2 A0A0P5BRP3 A0A0P5UMH7 A0A0P5IFM4 A0A0P5BRR9 A0A0P5Y6Z4 A0A0N8C6G6 A0A0N8B3V9 Q17PM3 A0A0P4Z1P6 A0A023EY38 A0A126Q9D5 A0A182H564 E9GT82 A0A1W4XCF9 K1R512 A0A0N8DHY8 A0A1B6EL67 A0A0P5XQE1 U5EY13 A0A0P5HPX3 A0A1S3IS83 A0A0P5G4Z2 V3ZN51 A0A0P5KVE5 A0A0N8D1M8 A0A0P5SXH0 A0A1B6ECM6 B0WIK3 A0A1Q3F1Q3 A0A0P5RCA0 A0A1Q3F1R2 A0A210Q8D8 A0A0P5IES6 A0A0P5RAE9 A0A2C9JNE9 A0A1Z5LGF5 A0A0P4ZBL9 A0A0P5AQQ8 A0A2R5LE81 A0A0P6G5Q7 A0A0P6EE22 A0A1L8DGR0 G1ARD5 A0A0P5K1F1 A0A0P5VBJ5 A0A0P5VEH1 A0A0B6ZVN1 A0A0N8CGJ0 A0A1J1IXN9 A0A2I9LPS2 A0A087ULG5 A0A0P5QNC1 A0A0P4XWZ0 A0A0P5IIZ5 A7RXU0 A0A1I8N1Y7 N1PBE4 A0A0P5R6M2 A0A0P4ZZC7 A0A0A1WIC5 A0A1W4VNR6 T1PB73 A0A0P4Z5X6 B4KXY0 B4MGQ8 B4QEY1 A0A0L8GAX7 A0A0M3QVT8 N6TSV1 Q9W0S7 A0A1I8PTZ6 B3NEM9
A0A2A4J4G7 A0A194QN47 A0A194PF40 A0A2J7RK75 A0A2J7RK72 A0A067RA05 A0A1L7NZM8 A0A1Y1NAS3 A0A0P5RPB4 A0A0P6ECC7 A0A0P6D5Y7 A0A0P5JM29 D6WH29 A0A0P5M5M6 A0A0P6DAC5 A0A0T6AYP2 A0A0P5NMF8 A0A0P5MWB4 A0A0P6DA35 A0A0P6F8J1 A0A0P6IWG6 A0A0P5VNB2 A0A0P5BRP3 A0A0P5UMH7 A0A0P5IFM4 A0A0P5BRR9 A0A0P5Y6Z4 A0A0N8C6G6 A0A0N8B3V9 Q17PM3 A0A0P4Z1P6 A0A023EY38 A0A126Q9D5 A0A182H564 E9GT82 A0A1W4XCF9 K1R512 A0A0N8DHY8 A0A1B6EL67 A0A0P5XQE1 U5EY13 A0A0P5HPX3 A0A1S3IS83 A0A0P5G4Z2 V3ZN51 A0A0P5KVE5 A0A0N8D1M8 A0A0P5SXH0 A0A1B6ECM6 B0WIK3 A0A1Q3F1Q3 A0A0P5RCA0 A0A1Q3F1R2 A0A210Q8D8 A0A0P5IES6 A0A0P5RAE9 A0A2C9JNE9 A0A1Z5LGF5 A0A0P4ZBL9 A0A0P5AQQ8 A0A2R5LE81 A0A0P6G5Q7 A0A0P6EE22 A0A1L8DGR0 G1ARD5 A0A0P5K1F1 A0A0P5VBJ5 A0A0P5VEH1 A0A0B6ZVN1 A0A0N8CGJ0 A0A1J1IXN9 A0A2I9LPS2 A0A087ULG5 A0A0P5QNC1 A0A0P4XWZ0 A0A0P5IIZ5 A7RXU0 A0A1I8N1Y7 N1PBE4 A0A0P5R6M2 A0A0P4ZZC7 A0A0A1WIC5 A0A1W4VNR6 T1PB73 A0A0P4Z5X6 B4KXY0 B4MGQ8 B4QEY1 A0A0L8GAX7 A0A0M3QVT8 N6TSV1 Q9W0S7 A0A1I8PTZ6 B3NEM9
PDB
3BDL
E-value=1.91182e-136,
Score=1248
Ontologies
GO
Topology
Subcellular location
Cytoplasm
Nucleus Associates with ribosomes. With evidence from 4 publications.
Nucleus Associates with ribosomes. With evidence from 4 publications.
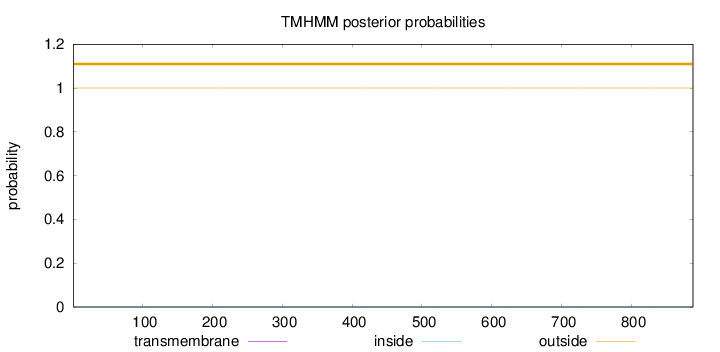
Length:
888
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00121
Exp number, first 60 AAs:
0.00077
Total prob of N-in:
0.00006
outside
1 - 888
Population Genetic Test Statistics
Pi
19.656589
Theta
16.2085
Tajima's D
0.40673
CLR
0.129873
CSRT
0.496075196240188
Interpretation
Uncertain
