Gene
KWMTBOMO00460 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA013317
Annotation
PREDICTED:_carnitine_O-palmitoyltransferase_1?_liver_isoform_[Papilio_machaon]
Location in the cell
PlasmaMembrane Reliability : 2.765
Sequence
CDS
ATGGCAGAAGCTCATTCGGCTGTGGCATTTTCCTTCGCAATAACTCACGATGGCTGGGATGTGAATTTCGACCGTGAAGTCCTCTACCTAGTTTGGGAATCGGGTGTTCGATCATGGAAGAAACGTCTTGCTCGTTTTAGGAACAATGTACTGAATGGCGTGTACCCAGGTCATCTGCAGTCACTTTATGTGCTGTGGACACTACTTGTAGCTGCCCATTTCAGTAACTTCAACATACCATTTGGCTTAGTTGATAAAATGATAACAATAATGCCAAGCAATTCAACATGGGCCCAGGTAGTAGCGTGCTTCTTAGTAGCGCTCACCATGTGGTTTATCGTTATATTTTTGATGAGATATCTGCTGAAATTACTGCTTATGTACAAGGGTTGGATGTATGAGTCACGTGGTCCCGGTGCTAAAATATCGTTAAAGACCAAACTGTGGGTGAGCGTCGTGAAAGTACTATCCGGATGGAACAAACCAAAGTTGTACAGTTTTCAAGGCTCTTTGCCCAGACTCCCTCTGCCGGCTGTTGAAGATACTATGAATAGGTACCTACTCAGTGTCCGACCATTACTTGACAATGATAATTACAAAAGAGTGGAAAAATTAGCTGTAGAATTCAAAACAAAAATTGCGGTTAAACTACAGAGATATCTGATACTCAAATCTTGGTGGTCTAGCAACTACGTATCGGACTGGTGGGAGGAGTATGTTTATTTAAAAGGTCGCTCACCCCTGATGGTCAATTCCAATTTCTATGGAACAGATGCTATTTTATTGAGACCTACACAGATACAAGCGGCCCGTGCGGGCGTCATCATCCACCTCTGCTTAAAGTTCCGTAGGCTTATCGATAGTCAAGAATTGGAACCCATAATGCTACAAAACATGGTTCCCCTGTGCTCCTGGCAATACGAGCGCCTTTTCAACACGGTACGCGCGCCTGGCATCGAAACCGATAAACTTGTGCACTATCAAGACTCCTCGCACATCGTGGTTTATCATAAAGGCAAATATTTTAAGGTGTTGGTTTACTTGAGGGGTCGTCTGTTAAAACCGGCAGAAATTCAAGTGCAAATACAACAGATACTGGACGATAAATCGGAGCCGGCGATCGGTGAGGAGAAACTGGCCGCACTAACCGCCGGAGAGCGAACGCATTGGGCCCAAATCCGACAGACGATGTTCAATCGCGGCCACAACCGTACATCCCTGCATTGCATTGAGCGCGCTGCCTTCACGGTCAGCTTGGACGATATGGAATATGGCTACGACGAGACCGATCCAAAGAAGCTGGATGAGTTTGGTCACGTGCTACTGCACGGTAAAGGATATGATCGTTGGTTCGACAAATCCTTCAACTTATGTTTTAGTACTAATGGCTTCGTTGGATTCAATGCGGAACACAGTTGGGCCGATGCGCCTGTTATGGGTCATCTTTGGGAATACGTTATTTGGTCTGAACTTGAAATAGGGTACCAGCCGGATGGCAACACCAAGGGCACTATTGTTACTAATCCGCCAGCGCCAATACGTCTCCAGTGGGAGTTAAATAAAGAAGAAATATTAAGCGCCATTGATATTTCGTATAGCGTCGCTCATAAACTCCTCAATGATGTAGACCTAAGGATTCTGATGTACACCAAATATGGAAAAGGTTTCATGAAGAAATGCCGCGTATCTCCGGACGCATTCATTCAGCTGATTCTCCAACTGTCTTACTACCGCGACGCAAGCCGTTTCTGTCTCACGTATGAGGCGTCTATGACCAGATTGTTCCGCGAAGGTCGCACGGAGACAGTAAGATCCTGCACCATTGAAAGCACCGAGTGGGTCCGCGCCATGGAAAATCCTAACAGTAGTATCGAAGAGCGCATGGCGTTATTCATAAAAGCCAGTAACAGACATCAGCTCGGCTACCAAGACGCTATGGCGGGTCGGGGCATCGACCGCCACCTTTTCTGTTTGTATGTCGTCGCTAAATATTTGGAACTCGAGTCGCCCTTCTTGCAAGAAGTCTTCAATGAACCATGGCGTCTGTCCACCAGCCAAACACCCTACGGTCAAACCAGCCTCCTGGATCTGAAAAAGTATCCTAAATGTGTAAGCGCGGGTGGTGGGTTCGGTCCCGTCGCGGATGACGGCTATGGTGTCTCGTACATCATTGTCGGTGAAGATACTATATTTTTCCACGTCTCAAGCAAGAAGAGCTGCCCAATAACCAGTTCAACGAACTTCGTGAAGCAAATCGAGAAATCCCTTCACGACGTTCAAACATTGTTTAATGATTACAACAAACTGAAGAAAACCAACAAACCGATAACAGACTAA
Protein
MAEAHSAVAFSFAITHDGWDVNFDREVLYLVWESGVRSWKKRLARFRNNVLNGVYPGHLQSLYVLWTLLVAAHFSNFNIPFGLVDKMITIMPSNSTWAQVVACFLVALTMWFIVIFLMRYLLKLLLMYKGWMYESRGPGAKISLKTKLWVSVVKVLSGWNKPKLYSFQGSLPRLPLPAVEDTMNRYLLSVRPLLDNDNYKRVEKLAVEFKTKIAVKLQRYLILKSWWSSNYVSDWWEEYVYLKGRSPLMVNSNFYGTDAILLRPTQIQAARAGVIIHLCLKFRRLIDSQELEPIMLQNMVPLCSWQYERLFNTVRAPGIETDKLVHYQDSSHIVVYHKGKYFKVLVYLRGRLLKPAEIQVQIQQILDDKSEPAIGEEKLAALTAGERTHWAQIRQTMFNRGHNRTSLHCIERAAFTVSLDDMEYGYDETDPKKLDEFGHVLLHGKGYDRWFDKSFNLCFSTNGFVGFNAEHSWADAPVMGHLWEYVIWSELEIGYQPDGNTKGTIVTNPPAPIRLQWELNKEEILSAIDISYSVAHKLLNDVDLRILMYTKYGKGFMKKCRVSPDAFIQLILQLSYYRDASRFCLTYEASMTRLFREGRTETVRSCTIESTEWVRAMENPNSSIEERMALFIKASNRHQLGYQDAMAGRGIDRHLFCLYVVAKYLELESPFLQEVFNEPWRLSTSQTPYGQTSLLDLKKYPKCVSAGGGFGPVADDGYGVSYIIVGEDTIFFHVSSKKSCPITSSTNFVKQIEKSLHDVQTLFNDYNKLKKTNKPITD
Summary
Uniprot
H9JUV4
A0A2W1BRW1
A0A2A4ISR3
A0A2H1W6J1
A0A1E1WMU0
A0A194QLF8
+ More
A0A194PGR8 A0A212EKH5 K7IW67 V5GW75 A0A1B6L5C7 D6WI25 A0A084VPZ2 A0A182K3P3 A0A1B6CIN1 A0A182QLA7 A0A2M3Z0Q8 A0A182UMD3 A0A067RA00 A0A2M4BEF0 A0A2J7RKA6 A0A182XB12 A0A2M4A0T4 A0A1S4H375 A0A182KJM4 A0A182I0K5 A0A182R7R4 A0A182N8N8 E2A6C2 A0A336M8Q5 A0A336M930 Q7Q0U0 A0A182M9R9 T1E1Y6 A0A1B6DCM8 A0A182FMY5 A0A0K8TTA0 A0A232FDJ0 A0A1I8M899 A0A026WET5 A0A182U978 A0A2J7RK93 A0A336KRP5 A0A336M4U0 A0A1Y1JVJ2 A0A1Q3FF05 A0A182YIL3 A0A336KMI8 A0A1Q3FEQ2 A0A1Q3FEP3 A0A336KLM4 A0A1Q3FEQ5 W5J7N7 A0A336KSM9 Q179Z5 A0A195CTF6 A0A336KKA7 A0A1Y1JSD4 A0A182IW25 T1PKH8 A0A023EW28 A0A151JZG1 A0A151XFQ3 A0A195BJC2 A0A158NFD0 B0WED0 F4WUJ2 A0A1I8NSM4 A0A034VPK8 A0A195E8G3 A0A0Q9W5H9 A0A023F564 A0A0L0BTS0 A0A2A3E9G3 A0A1L8EE34 B3MI43 Q7JQH9 A0A336KPU3 A0A336KLU3 D3TRL5 A0A1A9V5B4 B4P871 A0A0Q9XGI9 Q28XI5 A0A034VKD7 A0A1L8DYB6 A0A1B0AHR2 A0A0C9RVR9 A0A1L8DYE7 A0A0Q5VN39 A0A0J9R9P8 A0A1W4X321 A0A1I8NSM7 A0A0V0G650 A0A1A9WJ04 A0A0P9C0X8 A0A1W4VP57 A0A1W4X490 A0A146KJV7 A0A0K8U212
A0A194PGR8 A0A212EKH5 K7IW67 V5GW75 A0A1B6L5C7 D6WI25 A0A084VPZ2 A0A182K3P3 A0A1B6CIN1 A0A182QLA7 A0A2M3Z0Q8 A0A182UMD3 A0A067RA00 A0A2M4BEF0 A0A2J7RKA6 A0A182XB12 A0A2M4A0T4 A0A1S4H375 A0A182KJM4 A0A182I0K5 A0A182R7R4 A0A182N8N8 E2A6C2 A0A336M8Q5 A0A336M930 Q7Q0U0 A0A182M9R9 T1E1Y6 A0A1B6DCM8 A0A182FMY5 A0A0K8TTA0 A0A232FDJ0 A0A1I8M899 A0A026WET5 A0A182U978 A0A2J7RK93 A0A336KRP5 A0A336M4U0 A0A1Y1JVJ2 A0A1Q3FF05 A0A182YIL3 A0A336KMI8 A0A1Q3FEQ2 A0A1Q3FEP3 A0A336KLM4 A0A1Q3FEQ5 W5J7N7 A0A336KSM9 Q179Z5 A0A195CTF6 A0A336KKA7 A0A1Y1JSD4 A0A182IW25 T1PKH8 A0A023EW28 A0A151JZG1 A0A151XFQ3 A0A195BJC2 A0A158NFD0 B0WED0 F4WUJ2 A0A1I8NSM4 A0A034VPK8 A0A195E8G3 A0A0Q9W5H9 A0A023F564 A0A0L0BTS0 A0A2A3E9G3 A0A1L8EE34 B3MI43 Q7JQH9 A0A336KPU3 A0A336KLU3 D3TRL5 A0A1A9V5B4 B4P871 A0A0Q9XGI9 Q28XI5 A0A034VKD7 A0A1L8DYB6 A0A1B0AHR2 A0A0C9RVR9 A0A1L8DYE7 A0A0Q5VN39 A0A0J9R9P8 A0A1W4X321 A0A1I8NSM7 A0A0V0G650 A0A1A9WJ04 A0A0P9C0X8 A0A1W4VP57 A0A1W4X490 A0A146KJV7 A0A0K8U212
Pubmed
19121390
28756777
26354079
22118469
20075255
18362917
+ More
19820115 24438588 24845553 12364791 20966253 20798317 14747013 17210077 24330624 26369729 28648823 25315136 24508170 30249741 28004739 25244985 20920257 23761445 17510324 24945155 21347285 21719571 25348373 17994087 25474469 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20353571 17550304 15632085 18057021 22936249 26823975
19820115 24438588 24845553 12364791 20966253 20798317 14747013 17210077 24330624 26369729 28648823 25315136 24508170 30249741 28004739 25244985 20920257 23761445 17510324 24945155 21347285 21719571 25348373 17994087 25474469 26108605 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 20353571 17550304 15632085 18057021 22936249 26823975
EMBL
BABH01039784
KZ149977
PZC75897.1
NWSH01008073
PCG62709.1
ODYU01006633
+ More
SOQ48647.1 GDQN01002742 JAT88312.1 KQ461196 KPJ06398.1 KQ459605 KPI91899.1 AGBW02014274 OWR41999.1 GALX01003993 JAB64473.1 GEBQ01021080 JAT18897.1 KQ971334 EEZ99701.1 ATLV01015089 KE525002 KFB40036.1 GEDC01026748 GEDC01024028 JAS10550.1 JAS13270.1 AXCN02001857 GGFM01001366 MBW22117.1 KK852598 KDR20535.1 GGFJ01002288 MBW51429.1 NEVH01002981 PNF41260.1 GGFK01001096 MBW34417.1 AAAB01008980 APCN01005216 GL437123 EFN71007.1 UFQS01000675 UFQT01000675 SSX06074.1 SSX26430.1 UFQS01000558 UFQT01000558 SSX04931.1 SSX25293.1 EAA13810.3 AXCM01000376 GALA01001244 JAA93608.1 GEDC01013847 JAS23451.1 GDAI01000227 JAI17376.1 NNAY01000414 OXU28580.1 KK107260 QOIP01000001 EZA54166.1 RLU26627.1 PNF41258.1 SSX06075.1 SSX26431.1 SSX04930.1 SSX25292.1 GEZM01101960 JAV52161.1 GFDL01008911 JAV26134.1 SSX06076.1 SSX26432.1 GFDL01009019 JAV26026.1 GFDL01009018 JAV26027.1 SSX04932.1 SSX25294.1 GFDL01009015 JAV26030.1 ADMH02001931 ETN60467.1 SSX06073.1 SSX26429.1 CH477342 EAT43076.1 KQ977279 KYN03981.1 SSX04929.1 SSX25291.1 GEZM01101959 JAV52162.1 KA645126 KA649209 AFP63838.1 GAPW01000261 JAC13337.1 KQ981410 KYN41921.1 KQ982182 KYQ59187.1 KQ976464 KYM84546.1 ADTU01014263 DS231907 EDS45490.1 GL888365 EGI62157.1 GAKP01015225 GAKP01015222 JAC43730.1 KQ979440 KYN21505.1 CH940648 KRF80200.1 GBBI01002503 JAC16209.1 JRES01001351 KNC23442.1 KZ288323 PBC28114.1 GFDG01001873 JAV16926.1 CH902619 EDV38053.2 AE013599 BT010044 AAF58785.2 AAF58786.1 AAQ22513.1 SSX06072.1 SSX26428.1 SSX04928.1 SSX25290.1 CCAG010017066 EZ424067 ADD20343.1 CM000158 EDW90118.2 KRJ98945.1 CH933808 KRG04324.1 CM000071 EAL26331.3 GAKP01015223 GAKP01015221 JAC43731.1 GFDF01002643 JAV11441.1 GBYB01011796 JAG81563.1 GFDF01002630 JAV11454.1 CH954179 KQS62940.1 KQS62941.1 CM002911 KMY92798.1 KMY92800.1 GECL01002606 JAP03518.1 KPU77237.1 KPU77238.1 GDHC01021881 JAP96747.1 GDHF01031944 JAI20370.1
SOQ48647.1 GDQN01002742 JAT88312.1 KQ461196 KPJ06398.1 KQ459605 KPI91899.1 AGBW02014274 OWR41999.1 GALX01003993 JAB64473.1 GEBQ01021080 JAT18897.1 KQ971334 EEZ99701.1 ATLV01015089 KE525002 KFB40036.1 GEDC01026748 GEDC01024028 JAS10550.1 JAS13270.1 AXCN02001857 GGFM01001366 MBW22117.1 KK852598 KDR20535.1 GGFJ01002288 MBW51429.1 NEVH01002981 PNF41260.1 GGFK01001096 MBW34417.1 AAAB01008980 APCN01005216 GL437123 EFN71007.1 UFQS01000675 UFQT01000675 SSX06074.1 SSX26430.1 UFQS01000558 UFQT01000558 SSX04931.1 SSX25293.1 EAA13810.3 AXCM01000376 GALA01001244 JAA93608.1 GEDC01013847 JAS23451.1 GDAI01000227 JAI17376.1 NNAY01000414 OXU28580.1 KK107260 QOIP01000001 EZA54166.1 RLU26627.1 PNF41258.1 SSX06075.1 SSX26431.1 SSX04930.1 SSX25292.1 GEZM01101960 JAV52161.1 GFDL01008911 JAV26134.1 SSX06076.1 SSX26432.1 GFDL01009019 JAV26026.1 GFDL01009018 JAV26027.1 SSX04932.1 SSX25294.1 GFDL01009015 JAV26030.1 ADMH02001931 ETN60467.1 SSX06073.1 SSX26429.1 CH477342 EAT43076.1 KQ977279 KYN03981.1 SSX04929.1 SSX25291.1 GEZM01101959 JAV52162.1 KA645126 KA649209 AFP63838.1 GAPW01000261 JAC13337.1 KQ981410 KYN41921.1 KQ982182 KYQ59187.1 KQ976464 KYM84546.1 ADTU01014263 DS231907 EDS45490.1 GL888365 EGI62157.1 GAKP01015225 GAKP01015222 JAC43730.1 KQ979440 KYN21505.1 CH940648 KRF80200.1 GBBI01002503 JAC16209.1 JRES01001351 KNC23442.1 KZ288323 PBC28114.1 GFDG01001873 JAV16926.1 CH902619 EDV38053.2 AE013599 BT010044 AAF58785.2 AAF58786.1 AAQ22513.1 SSX06072.1 SSX26428.1 SSX04928.1 SSX25290.1 CCAG010017066 EZ424067 ADD20343.1 CM000158 EDW90118.2 KRJ98945.1 CH933808 KRG04324.1 CM000071 EAL26331.3 GAKP01015223 GAKP01015221 JAC43731.1 GFDF01002643 JAV11441.1 GBYB01011796 JAG81563.1 GFDF01002630 JAV11454.1 CH954179 KQS62940.1 KQS62941.1 CM002911 KMY92798.1 KMY92800.1 GECL01002606 JAP03518.1 KPU77237.1 KPU77238.1 GDHC01021881 JAP96747.1 GDHF01031944 JAI20370.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000053268
UP000007151
UP000002358
+ More
UP000007266 UP000030765 UP000075881 UP000075886 UP000075903 UP000027135 UP000235965 UP000076407 UP000075882 UP000075840 UP000075900 UP000075884 UP000000311 UP000007062 UP000075883 UP000069272 UP000215335 UP000095301 UP000053097 UP000279307 UP000075902 UP000076408 UP000000673 UP000008820 UP000078542 UP000075880 UP000078541 UP000075809 UP000078540 UP000005205 UP000002320 UP000007755 UP000095300 UP000078492 UP000008792 UP000037069 UP000242457 UP000007801 UP000000803 UP000092444 UP000078200 UP000002282 UP000009192 UP000001819 UP000092445 UP000008711 UP000192223 UP000091820 UP000192221
UP000007266 UP000030765 UP000075881 UP000075886 UP000075903 UP000027135 UP000235965 UP000076407 UP000075882 UP000075840 UP000075900 UP000075884 UP000000311 UP000007062 UP000075883 UP000069272 UP000215335 UP000095301 UP000053097 UP000279307 UP000075902 UP000076408 UP000000673 UP000008820 UP000078542 UP000075880 UP000078541 UP000075809 UP000078540 UP000005205 UP000002320 UP000007755 UP000095300 UP000078492 UP000008792 UP000037069 UP000242457 UP000007801 UP000000803 UP000092444 UP000078200 UP000002282 UP000009192 UP000001819 UP000092445 UP000008711 UP000192223 UP000091820 UP000192221
Interpro
SUPFAM
SSF52540
SSF52540
Gene 3D
ProteinModelPortal
H9JUV4
A0A2W1BRW1
A0A2A4ISR3
A0A2H1W6J1
A0A1E1WMU0
A0A194QLF8
+ More
A0A194PGR8 A0A212EKH5 K7IW67 V5GW75 A0A1B6L5C7 D6WI25 A0A084VPZ2 A0A182K3P3 A0A1B6CIN1 A0A182QLA7 A0A2M3Z0Q8 A0A182UMD3 A0A067RA00 A0A2M4BEF0 A0A2J7RKA6 A0A182XB12 A0A2M4A0T4 A0A1S4H375 A0A182KJM4 A0A182I0K5 A0A182R7R4 A0A182N8N8 E2A6C2 A0A336M8Q5 A0A336M930 Q7Q0U0 A0A182M9R9 T1E1Y6 A0A1B6DCM8 A0A182FMY5 A0A0K8TTA0 A0A232FDJ0 A0A1I8M899 A0A026WET5 A0A182U978 A0A2J7RK93 A0A336KRP5 A0A336M4U0 A0A1Y1JVJ2 A0A1Q3FF05 A0A182YIL3 A0A336KMI8 A0A1Q3FEQ2 A0A1Q3FEP3 A0A336KLM4 A0A1Q3FEQ5 W5J7N7 A0A336KSM9 Q179Z5 A0A195CTF6 A0A336KKA7 A0A1Y1JSD4 A0A182IW25 T1PKH8 A0A023EW28 A0A151JZG1 A0A151XFQ3 A0A195BJC2 A0A158NFD0 B0WED0 F4WUJ2 A0A1I8NSM4 A0A034VPK8 A0A195E8G3 A0A0Q9W5H9 A0A023F564 A0A0L0BTS0 A0A2A3E9G3 A0A1L8EE34 B3MI43 Q7JQH9 A0A336KPU3 A0A336KLU3 D3TRL5 A0A1A9V5B4 B4P871 A0A0Q9XGI9 Q28XI5 A0A034VKD7 A0A1L8DYB6 A0A1B0AHR2 A0A0C9RVR9 A0A1L8DYE7 A0A0Q5VN39 A0A0J9R9P8 A0A1W4X321 A0A1I8NSM7 A0A0V0G650 A0A1A9WJ04 A0A0P9C0X8 A0A1W4VP57 A0A1W4X490 A0A146KJV7 A0A0K8U212
A0A194PGR8 A0A212EKH5 K7IW67 V5GW75 A0A1B6L5C7 D6WI25 A0A084VPZ2 A0A182K3P3 A0A1B6CIN1 A0A182QLA7 A0A2M3Z0Q8 A0A182UMD3 A0A067RA00 A0A2M4BEF0 A0A2J7RKA6 A0A182XB12 A0A2M4A0T4 A0A1S4H375 A0A182KJM4 A0A182I0K5 A0A182R7R4 A0A182N8N8 E2A6C2 A0A336M8Q5 A0A336M930 Q7Q0U0 A0A182M9R9 T1E1Y6 A0A1B6DCM8 A0A182FMY5 A0A0K8TTA0 A0A232FDJ0 A0A1I8M899 A0A026WET5 A0A182U978 A0A2J7RK93 A0A336KRP5 A0A336M4U0 A0A1Y1JVJ2 A0A1Q3FF05 A0A182YIL3 A0A336KMI8 A0A1Q3FEQ2 A0A1Q3FEP3 A0A336KLM4 A0A1Q3FEQ5 W5J7N7 A0A336KSM9 Q179Z5 A0A195CTF6 A0A336KKA7 A0A1Y1JSD4 A0A182IW25 T1PKH8 A0A023EW28 A0A151JZG1 A0A151XFQ3 A0A195BJC2 A0A158NFD0 B0WED0 F4WUJ2 A0A1I8NSM4 A0A034VPK8 A0A195E8G3 A0A0Q9W5H9 A0A023F564 A0A0L0BTS0 A0A2A3E9G3 A0A1L8EE34 B3MI43 Q7JQH9 A0A336KPU3 A0A336KLU3 D3TRL5 A0A1A9V5B4 B4P871 A0A0Q9XGI9 Q28XI5 A0A034VKD7 A0A1L8DYB6 A0A1B0AHR2 A0A0C9RVR9 A0A1L8DYE7 A0A0Q5VN39 A0A0J9R9P8 A0A1W4X321 A0A1I8NSM7 A0A0V0G650 A0A1A9WJ04 A0A0P9C0X8 A0A1W4VP57 A0A1W4X490 A0A146KJV7 A0A0K8U212
PDB
1XMC
E-value=3.30783e-76,
Score=728
Ontologies
PATHWAY
GO
PANTHER
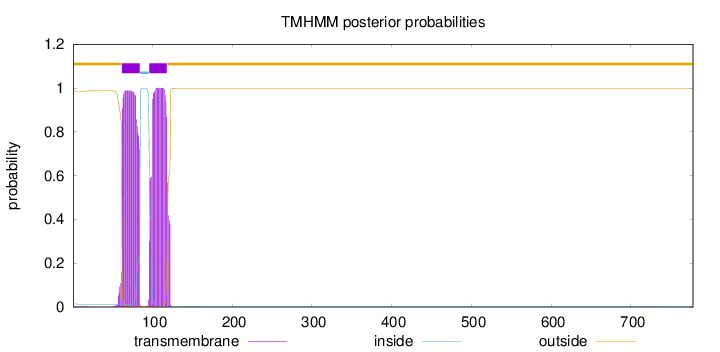
Topology
Length:
778
Number of predicted TMHs:
2
Exp number of AAs in TMHs:
44.7043500000001
Exp number, first 60 AAs:
0.38814
Total prob of N-in:
0.01408
outside
1 - 61
TMhelix
62 - 84
inside
85 - 95
TMhelix
96 - 118
outside
119 - 778
Population Genetic Test Statistics
Pi
14.967614
Theta
18.531631
Tajima's D
-2.232577
CLR
0.072298
CSRT
0.00384980750962452
Interpretation
Uncertain
