Gene
KWMTBOMO00456
Pre Gene Modal
BGIBMGA003354
Annotation
PREDICTED:_uncharacterized_protein_LOC106720810_[Papilio_machaon]
Full name
ATP-dependent DNA helicase
Location in the cell
Nuclear Reliability : 3.215
Sequence
CDS
ATGTCGTCGACCGAGTCGGAGTCTGTTTCAAATGCAGAGTACGAAAGTTTATCCCAGAAAATCATCGTGGAAGTACAGAAAAGACCAGCTTTATACGATACTACTTTGCGTCAATACAGTGATAGAAATGTGAAAAATAAACTTTGGGAGGAAGTTTATCACAACGTAGTTAATAATTGGATGACACTGTCGGAGGCTGAGAAAAAAAAGAAGGGTAGTACAATACAAAAGAAATGGAAGAATATGCGCGACAGTTTTTCTAAAGAATTATACAGCGTGGTAGTTAAGGTAGCTGTTAGATGTAGACTCAGGCCCGTCAGACCCTGGATGCTGAACGCCACGCATCTCAGAGCTGCTGAGACAGTGGAACAGACTCAGACACGTCGAGTTTTAGATGCCGAACGTCACGCGCAATTGATTGCGGCAGAGACTGACGAAGATTCACAAAGACGACGTCTTTTAAATGCTGAACGGCAGGCAGAAAGAAGACGTACGTTTTCAATGAGTACGTGGGAGGCATTTAATGGTGCCGCTTTTGAGTATGACCCTTTGATCGACTATGTTAATCACCGATTGGTTATCATTGGGAGAATGGATAAAAAATGCAGATATTGTGGAGCACTGAAATGGAAAGAAGAAACTCCAGGTATGTGCTGTTCTGGTGGAAAGGTTAAAATTCCATTACTTGGCGAGCCAGAGGAACCTCTTAAATCTTTACTTTTATACGACAGTAACGAATCGCGCCGGTTTTTAAGCAGGATTAGAAAGTATAATTCTTGCTTTCAGATGACGTCATTTGGTGTAGATAAGGAGGTCGTAATGCCTGGATTTTCACCTACTTTTACCATCCAAGGACAGGTGTATCACAGGATTGGTTCCTTACTTCCCGCAAATAATGAACAACCAAAGCTTTTGCAACTTTATTTCATGGGCGATGAAAAAAACGAAGCTGATCGCAGGTGTCAAAATATACAAGGAATCGAAAGGGATACTGTTTTAAAAATCCAGAGAATGTTGCATGAACATAATCGCCTTATCAATACTTTCAAAACAGCACTAGAAAGAATGCCGGGAGAGGATTATAGGTTGGTGATGCACCCCGATCGCACACCAAGTGGGGAACATGAAAGGCGGTATAATGCACCGTTAATAAATGAAGTCGCAGCCTTGGTTACAGGCGAACAGTTTGCTTTCCGTGATATAGTTTTACAGGCCCGAGATGACACATTAACCAGGGTGCCTGATACTCATAAATTTTATGATGCGTTGCAATATCCGATCATATTTAGTAAGGGACAAGAGGGGTACCACTTTCAAGTTCCTCAAATTAATCCTGTAACAGGTCTTCCCTTGCATAACAAAAAGGTTTCATGCATGGATTTTTATGCCTACAACATGATGATACGAGAAAACAACTTTAACATTGTACAAAGATGCAAGCAACTGGCCAATCAGTCTTACGTTGACATGTACGTTAAAGTTGAAAGCGAGAGGTTACGGTACATATCATTAAATCAAACTAAACTAAGAGCAGAGAATTACATCCATCTTCAAGACGCTGTGGCAAATGACGCTAATTTAAATCCAAATAACTTAGGTCGTATGGTCATTTTACCATCTTCATTTGTAAATAGCCCGCGGTACTTACACGAATATACTCAGGACGCGTTTGTTTATGTGCGTACACATGGTCGCCCTGACTTATTCGTCACCTTTACTTGCAACCCAGCCTGGCCCGAAATAGTCAATCAGTTGATGCCGGGGCAAAGCGCAATAGATAGACATGATGTAGTCGCAAGAGTTTTGAGACTAAAAGTTAAAAAACTAATGGATGTGATTAATAAAGGCCGAGTGTTCAGAGAAGCGCGCTGCTTTATGTACTCTGTCGAATGGCAGAAACGTGGACTGCCACATGTCCATATATTATTGTGGTTAAAAGATAAGTTACGACCCGATCAAATAGATAACATAATCAGCGCTGAAATACCGGATCCTAACATTGACAAGACTCTCCATGACATTATTGTAAAAAATATGATTCACGGTCCATGCGGACCCGAAAATCCACAGTGCCACTGCATGAAAGACGGAAAATGCACAAAAAAATTTCCTCGCAAGTTAGTGAAAGAGACCGTTCACAACGACCACGGATACCCGCAGTATCGTAGAAGAGCGCCAGCAGACGGAGGTCGAACAGCAACCGTGAAGCTCCGAAATGGTAGCTACGTTACAGTTGATAATAGTTGGGTAGTCCCCTATTCACCAATTTTATCAAAAATGTTTAACGCCCACATAAATGTTGAGGCATGCAGTTCAGTACGCGCCATCAAATATATTTGCAAATATATAAACAAGGGTAGTGATCAAGCTATTTTCAATTTTAGAAATACTGATCTTGCGAATCCCGTAGATGAGGTACAAACTTATCAGTCTGGTCGGTATGTCAGCAGCAACGAAGCCGTGTGGCGTCTGTTAGCATTTCCGTTGCACGAAAGGCATCCCACCGTGACACATCTCAGCGTGCATTTAGAAAATGGTGAACGCGTTTATTTTAATGAACACAATTTCCACGAACGAATAACTACTCCGCCAAAAACCACACTCACTGCTTTTTTTGATCTTTGTATCAGAGATGAGTTTGCAAAAACTCTTCTTTATGTCGAGGTGCCGAGGTATTATACTTGGGATGCAAGTAGGAAAACTTGGAAACGCCGCATTCAAGGTACTTCAGTTCACGATTGGCCTGGCGTCAAATCTGGAGATGCTTTAGGACGAGTTTATACAGTTCATGTTAGTAACATGGAATGTTTTTGTCTACGCATGCTTTTACACCATGTGCGCGGACCTACCTCTTTCAATGATCTAAAAAAATACAACACACAAGATTATCCTACATTTCGAGAGGCATGTGAGGCAAGAGGATTACTGGAGAATGACAACCATTGGGATGTGACACTGGAAGAAGCTGCTCAATGTAGATCAGCAGGCAAGATGAGAGAGCTATTTGCTGTATTAGTAGCGACATGTGGTCTTTCAAATTCTCAACAATTGTGGGATAAATACAAAAATGATATGACCGACGATATATTGCACAGATTACAAGAGCAAAATCCCAATGTCACGTACAGTGATTTAATTTACAATGAGGGTCTGACAAAAATTGAAAACCAGGTTAAAACAATTTCTAGAAAGGATTTATCAGATTATGGAATGATCAGACCACAGAGAACCGAGGAAATCAGTAGCGATTTAATACGGGAACTAGATTACGATACTGCTTCCTTACAACAACAATTGACAGAGTCTGTTCCACGTTTGATCCCAGAACAAAGGTTAGTATTTGACAATGTTTTCAAAAAAATCGAAGATGGCGAAGGTGGTTTGTTATTTTTGGATGCTCCCGGAGGCACGGGAAAAACCTTTCTTTTAAACTTGCTTTTAGCGCAAATCCGAAAAGATAAAGGAATTGCTGTAGCAGTTGCCTCTTCCGGAATAGCCGCCACTCTGCTCAATGGTGGTCGAACGGCACATTCGGTGCTTAAACTGTCATTAAACTTAGCCCATGAAGAAATGCCAGTCTGTAATATAAGTAAAAACAGCGAGCGTGGACGTATGTTGCAGCAATGTAAATTATTAGTTTGGGATGAGTGTACTATGTCCCATAAAAGAGCAATTGAAGCTCTAGATCGGACTTTGAAAGACATCAAGAGTAATCCAAATATCATGGGGGGGGATGGTGGTACTTTTAGCGGGCGATTTTAG
Protein
MSSTESESVSNAEYESLSQKIIVEVQKRPALYDTTLRQYSDRNVKNKLWEEVYHNVVNNWMTLSEAEKKKKGSTIQKKWKNMRDSFSKELYSVVVKVAVRCRLRPVRPWMLNATHLRAAETVEQTQTRRVLDAERHAQLIAAETDEDSQRRRLLNAERQAERRRTFSMSTWEAFNGAAFEYDPLIDYVNHRLVIIGRMDKKCRYCGALKWKEETPGMCCSGGKVKIPLLGEPEEPLKSLLLYDSNESRRFLSRIRKYNSCFQMTSFGVDKEVVMPGFSPTFTIQGQVYHRIGSLLPANNEQPKLLQLYFMGDEKNEADRRCQNIQGIERDTVLKIQRMLHEHNRLINTFKTALERMPGEDYRLVMHPDRTPSGEHERRYNAPLINEVAALVTGEQFAFRDIVLQARDDTLTRVPDTHKFYDALQYPIIFSKGQEGYHFQVPQINPVTGLPLHNKKVSCMDFYAYNMMIRENNFNIVQRCKQLANQSYVDMYVKVESERLRYISLNQTKLRAENYIHLQDAVANDANLNPNNLGRMVILPSSFVNSPRYLHEYTQDAFVYVRTHGRPDLFVTFTCNPAWPEIVNQLMPGQSAIDRHDVVARVLRLKVKKLMDVINKGRVFREARCFMYSVEWQKRGLPHVHILLWLKDKLRPDQIDNIISAEIPDPNIDKTLHDIIVKNMIHGPCGPENPQCHCMKDGKCTKKFPRKLVKETVHNDHGYPQYRRRAPADGGRTATVKLRNGSYVTVDNSWVVPYSPILSKMFNAHINVEACSSVRAIKYICKYINKGSDQAIFNFRNTDLANPVDEVQTYQSGRYVSSNEAVWRLLAFPLHERHPTVTHLSVHLENGERVYFNEHNFHERITTPPKTTLTAFFDLCIRDEFAKTLLYVEVPRYYTWDASRKTWKRRIQGTSVHDWPGVKSGDALGRVYTVHVSNMECFCLRMLLHHVRGPTSFNDLKKYNTQDYPTFREACEARGLLENDNHWDVTLEEAAQCRSAGKMRELFAVLVATCGLSNSQQLWDKYKNDMTDDILHRLQEQNPNVTYSDLIYNEGLTKIENQVKTISRKDLSDYGMIRPQRTEEISSDLIRELDYDTASLQQQLTESVPRLIPEQRLVFDNVFKKIEDGEGGLLFLDAPGGTGKTFLLNLLLAQIRKDKGIAVAVASSGIAATLLNGGRTAHSVLKLSLNLAHEEMPVCNISKNSERGRMLQQCKLLVWDECTMSHKRAIEALDRTLKDIKSNPNIMGGDGGTFSGRF
Summary
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Cofactor
Mg(2+)
Similarity
Belongs to the helicase family.
Feature
chain ATP-dependent DNA helicase
Uniprot
A0A267GN76
A0A267GEW9
A0A267F4D8
A0A267GWL2
A0A0D6L4E4
A0A016W7D9
+ More
A0A267F5E2 A0A0C2G6C2 A0A267F2R0 A0A1S3DI71 F4ZH17 A0A267FMY4 A0A0C2CWG5 E2AZ86 X1WX02 J9LB24 A0A368GX53 X1X109 A0A267F6F6 X1WRV4 A0A267EIS0 A0A267G464 A0A267ELF5 A0A267GG99 A0A267ETE1 A0A267DCD1 A0A1I8JBS8 A0A2H1W9S9 A0A267H554 A0A267ESS5 A0A2Z6RD64 A0A2H5SHM7 A0A2H5R2Y1 A0A015II56 A0A369KA40 A0A1R3JQ70 A0A2H5T4Y4 A8NPE0 A0A2H5U831 A0A2N9E605 A0A1R3GLH3 A0A397KYL5 Q5NAA4 Q0JP44 A0A0C2WEL5 A0A1Q3ENJ5 A0A267E9Q5 A0A267EKI4 Q7G4T8 Q8LML8 A0A0D2ZUX3 Q0IYM1 Q7XG88 Q8H878 A0A146IJN4 A0A369K0A0 A0A2N9I2F9 A0A2N9IBN4 A0A1D6IHV6 A0A1D6JV61 A0A1D6JX28 A0A3L6DGF1 A0A3L6FXZ4 A0A1D6IDS8 A0A317Y9Z1 A0A3L6E0C9 A0A1D6QRU3 A0A1D6P1V6 A0A3L6DAD7
A0A267F5E2 A0A0C2G6C2 A0A267F2R0 A0A1S3DI71 F4ZH17 A0A267FMY4 A0A0C2CWG5 E2AZ86 X1WX02 J9LB24 A0A368GX53 X1X109 A0A267F6F6 X1WRV4 A0A267EIS0 A0A267G464 A0A267ELF5 A0A267GG99 A0A267ETE1 A0A267DCD1 A0A1I8JBS8 A0A2H1W9S9 A0A267H554 A0A267ESS5 A0A2Z6RD64 A0A2H5SHM7 A0A2H5R2Y1 A0A015II56 A0A369KA40 A0A1R3JQ70 A0A2H5T4Y4 A8NPE0 A0A2H5U831 A0A2N9E605 A0A1R3GLH3 A0A397KYL5 Q5NAA4 Q0JP44 A0A0C2WEL5 A0A1Q3ENJ5 A0A267E9Q5 A0A267EKI4 Q7G4T8 Q8LML8 A0A0D2ZUX3 Q0IYM1 Q7XG88 Q8H878 A0A146IJN4 A0A369K0A0 A0A2N9I2F9 A0A2N9IBN4 A0A1D6IHV6 A0A1D6JV61 A0A1D6JX28 A0A3L6DGF1 A0A3L6FXZ4 A0A1D6IDS8 A0A317Y9Z1 A0A3L6E0C9 A0A1D6QRU3 A0A1D6P1V6 A0A3L6DAD7
EC Number
3.6.4.12
Pubmed
EMBL
NIVC01000229
PAA87485.1
NIVC01000366
PAA84583.1
NIVC01001445
PAA67902.1
+ More
NIVC01000111 PAA90396.1 KE126587 EPB66060.1 JARK01000714 EYC35207.1 NIVC01001359 PAA68995.1 KN743176 KIH52626.1 NIVC01001486 PAA67379.1 HQ009558 AEE09607.1 NIVC01000900 PAA75136.1 KN739487 KIH54222.1 GL444143 EFN61253.1 ABLF02041819 ABLF02018127 JOJR01000041 RCN48933.1 ABLF02020814 ABLF02049665 NIVC01001383 PAA68602.1 ABLF02017703 ABLF02041883 NIVC01002034 PAA61410.1 NIVC01000566 PAA80831.1 NIVC01001945 PAA62370.1 NIVC01000381 PAA84309.1 NIVC01001722 PAA64736.1 NIVC01004666 PAA46875.1 ODYU01007113 SOQ49602.1 NIVC01000042 PAA92669.1 NIVC01002018 NIVC01001743 NIVC01000051 PAA61561.1 PAA64516.1 PAA92464.1 BEXD01001738 GBB95559.1 BDIQ01000091 GBC29771.1 BDIQ01000010 GBC12419.1 JEMT01027503 EXX56857.1 LUEZ02000010 RDB29535.1 AWUE01015533 OMO96975.1 BDIQ01000117 GBC37632.1 AACS02000012 EAU86521.2 BDIQ01000188 GBC51003.1 OIVN01000105 SPC74366.1 AWWV01014072 OMO58887.1 KZ866277 RIA04643.1 AP003196 BAD81603.1 AP008207 BAF04484.1 KN818726 KIL54493.1 BDGU01000723 GAW08762.1 NIVC01002407 PAA58126.1 NIVC01002028 PAA61494.1 DP000086 AAP52492.2 AC105746 AAM92800.1 AP008216 BAF26194.2 AAP52578.2 AC104322 AAN09850.1 DF849855 GAT59324.1 LUEZ02000013 RDB28029.1 OIVN01004665 SPD18618.1 OIVN01005182 SPD21271.1 CM007650 ONM59070.1 CM007647 ONL95700.1 ONL96266.1 NCVQ01000010 PWZ07722.1 NCVQ01000003 PWZ39636.1 ONM57938.1 NCVQ01000001 PWZ54641.1 NCVQ01000008 PWZ14360.1 CM000780 AQK60210.1 CM000785 AQL04010.1 PWZ05017.1
NIVC01000111 PAA90396.1 KE126587 EPB66060.1 JARK01000714 EYC35207.1 NIVC01001359 PAA68995.1 KN743176 KIH52626.1 NIVC01001486 PAA67379.1 HQ009558 AEE09607.1 NIVC01000900 PAA75136.1 KN739487 KIH54222.1 GL444143 EFN61253.1 ABLF02041819 ABLF02018127 JOJR01000041 RCN48933.1 ABLF02020814 ABLF02049665 NIVC01001383 PAA68602.1 ABLF02017703 ABLF02041883 NIVC01002034 PAA61410.1 NIVC01000566 PAA80831.1 NIVC01001945 PAA62370.1 NIVC01000381 PAA84309.1 NIVC01001722 PAA64736.1 NIVC01004666 PAA46875.1 ODYU01007113 SOQ49602.1 NIVC01000042 PAA92669.1 NIVC01002018 NIVC01001743 NIVC01000051 PAA61561.1 PAA64516.1 PAA92464.1 BEXD01001738 GBB95559.1 BDIQ01000091 GBC29771.1 BDIQ01000010 GBC12419.1 JEMT01027503 EXX56857.1 LUEZ02000010 RDB29535.1 AWUE01015533 OMO96975.1 BDIQ01000117 GBC37632.1 AACS02000012 EAU86521.2 BDIQ01000188 GBC51003.1 OIVN01000105 SPC74366.1 AWWV01014072 OMO58887.1 KZ866277 RIA04643.1 AP003196 BAD81603.1 AP008207 BAF04484.1 KN818726 KIL54493.1 BDGU01000723 GAW08762.1 NIVC01002407 PAA58126.1 NIVC01002028 PAA61494.1 DP000086 AAP52492.2 AC105746 AAM92800.1 AP008216 BAF26194.2 AAP52578.2 AC104322 AAN09850.1 DF849855 GAT59324.1 LUEZ02000013 RDB28029.1 OIVN01004665 SPD18618.1 OIVN01005182 SPD21271.1 CM007650 ONM59070.1 CM007647 ONL95700.1 ONL96266.1 NCVQ01000010 PWZ07722.1 NCVQ01000003 PWZ39636.1 ONM57938.1 NCVQ01000001 PWZ54641.1 NCVQ01000008 PWZ14360.1 CM000780 AQK60210.1 CM000785 AQL04010.1 PWZ05017.1
Proteomes
Pfam
Interpro
IPR027417
P-loop_NTPase
+ More
IPR010285 DNA_helicase_pif1-like
IPR025476 Helitron_helicase-like
IPR038765 Papain-like_cys_pep_sf
IPR003653 Peptidase_C48_C
IPR003840 DNA_helicase
IPR003323 OTU_dom
IPR011990 TPR-like_helical_dom_sf
IPR002885 Pentatricopeptide_repeat
IPR013955 Rep_factor-A_C
IPR012340 NA-bd_OB-fold
IPR008254 Flavodoxin/NO_synth
IPR005062 SAC3/GANP/THP3
IPR018467 CCT_CS
IPR029039 Flavoprotein-like_sf
IPR010285 DNA_helicase_pif1-like
IPR025476 Helitron_helicase-like
IPR038765 Papain-like_cys_pep_sf
IPR003653 Peptidase_C48_C
IPR003840 DNA_helicase
IPR003323 OTU_dom
IPR011990 TPR-like_helical_dom_sf
IPR002885 Pentatricopeptide_repeat
IPR013955 Rep_factor-A_C
IPR012340 NA-bd_OB-fold
IPR008254 Flavodoxin/NO_synth
IPR005062 SAC3/GANP/THP3
IPR018467 CCT_CS
IPR029039 Flavoprotein-like_sf
Gene 3D
ProteinModelPortal
A0A267GN76
A0A267GEW9
A0A267F4D8
A0A267GWL2
A0A0D6L4E4
A0A016W7D9
+ More
A0A267F5E2 A0A0C2G6C2 A0A267F2R0 A0A1S3DI71 F4ZH17 A0A267FMY4 A0A0C2CWG5 E2AZ86 X1WX02 J9LB24 A0A368GX53 X1X109 A0A267F6F6 X1WRV4 A0A267EIS0 A0A267G464 A0A267ELF5 A0A267GG99 A0A267ETE1 A0A267DCD1 A0A1I8JBS8 A0A2H1W9S9 A0A267H554 A0A267ESS5 A0A2Z6RD64 A0A2H5SHM7 A0A2H5R2Y1 A0A015II56 A0A369KA40 A0A1R3JQ70 A0A2H5T4Y4 A8NPE0 A0A2H5U831 A0A2N9E605 A0A1R3GLH3 A0A397KYL5 Q5NAA4 Q0JP44 A0A0C2WEL5 A0A1Q3ENJ5 A0A267E9Q5 A0A267EKI4 Q7G4T8 Q8LML8 A0A0D2ZUX3 Q0IYM1 Q7XG88 Q8H878 A0A146IJN4 A0A369K0A0 A0A2N9I2F9 A0A2N9IBN4 A0A1D6IHV6 A0A1D6JV61 A0A1D6JX28 A0A3L6DGF1 A0A3L6FXZ4 A0A1D6IDS8 A0A317Y9Z1 A0A3L6E0C9 A0A1D6QRU3 A0A1D6P1V6 A0A3L6DAD7
A0A267F5E2 A0A0C2G6C2 A0A267F2R0 A0A1S3DI71 F4ZH17 A0A267FMY4 A0A0C2CWG5 E2AZ86 X1WX02 J9LB24 A0A368GX53 X1X109 A0A267F6F6 X1WRV4 A0A267EIS0 A0A267G464 A0A267ELF5 A0A267GG99 A0A267ETE1 A0A267DCD1 A0A1I8JBS8 A0A2H1W9S9 A0A267H554 A0A267ESS5 A0A2Z6RD64 A0A2H5SHM7 A0A2H5R2Y1 A0A015II56 A0A369KA40 A0A1R3JQ70 A0A2H5T4Y4 A8NPE0 A0A2H5U831 A0A2N9E605 A0A1R3GLH3 A0A397KYL5 Q5NAA4 Q0JP44 A0A0C2WEL5 A0A1Q3ENJ5 A0A267E9Q5 A0A267EKI4 Q7G4T8 Q8LML8 A0A0D2ZUX3 Q0IYM1 Q7XG88 Q8H878 A0A146IJN4 A0A369K0A0 A0A2N9I2F9 A0A2N9IBN4 A0A1D6IHV6 A0A1D6JV61 A0A1D6JX28 A0A3L6DGF1 A0A3L6FXZ4 A0A1D6IDS8 A0A317Y9Z1 A0A3L6E0C9 A0A1D6QRU3 A0A1D6P1V6 A0A3L6DAD7
Ontologies
GO
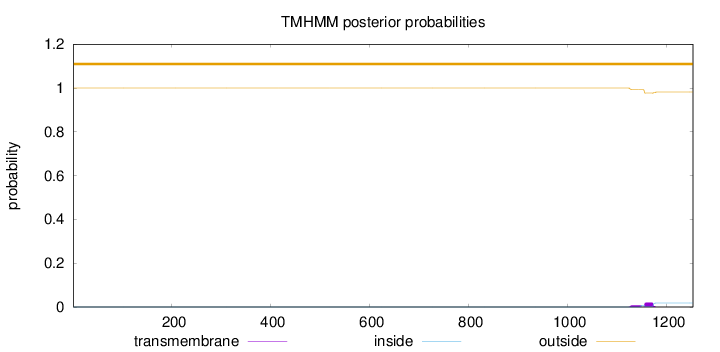
Topology
Length:
1253
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.5547
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00003
outside
1 - 1253
Population Genetic Test Statistics
Pi
15.553067
Theta
21.766281
Tajima's D
-1.553393
CLR
0
CSRT
0.0533473326333683
Interpretation
Uncertain
