Gene
KWMTBOMO00454
Annotation
PREDICTED:_uncharacterized_protein_LOC101738169_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 1.616 PlasmaMembrane Reliability : 2.09
Sequence
CDS
ATGACGCGATCTTCATTGTCTTATAAATGTGATTGTATTTACAGATTTAACCTTATTGCAATTGAGGATTTGCCCGCTGGGGAACCGATAGAGCAGGCATTGACGGAGCGAAGTAAGCGTTCATGGAACCCCCGAAATTATGAACCTGACGGAGAGGACATTCCAGTAATTTTGTCTTCACCTGCGATTCCGCCATCACCGGCAATGTACGTTGAATGTGGCGGCAGTGGTACGAGCACGCCATCTTCTAGATATAATTACAGATTTCTGTACATGGCAGCCGTAGTCCTTTGCCGTCACATAAACGTGTGCGTCGATCGCTTTGCCCAGACCAAGAACATGATACTACGAACAGAGATTCTATTTCATCGCTACGTCGACCGCTACGAGACTGAAATTTGCTACTTGTTGCACTATGGAAGTGACGACGATGACAGCGATGATGAGAGCCAGAATGATAACGACGAAATCGTTCCTATGATCATCCCCCGGGCAGCTGGAATACTGATGGACACTCCGGACGACGAGGAGATTGAGATTGACACGGCGGCGGCAAGTAAACCTCTTGTGACATCTTCCTCGCCTTCTATCATTTCAGCACCTACATCACCAGCGCGTGATACCGGTATGAAGCCTAGTCATTTTTTTGAATTTGACTGGGGAACTTTCCCTGACTCTCCTATACCACCAACAGAAAGGCGTGAATCTTTTAAGGAAAACTCTGGTCCCACGGTCACTGAGTACATGGAATTTATTGTGCAGGAAACAAATAGATATGCCCAACAATTAGCTGCTGAAATGTTGGACGGCGGGGAATTACAGGCCTCCAGTAGGATTACAGAATGGAAAGAAACCAATGTAGACGAGTTATTGGTGTTCTTCGGAATATTACTGGCGATGGGAATCGTCATAAAGAACCGGGTTGAAGAGTACTGGAATACGGAACAAAACATCTTCTCCACTCCAGGTTTCAAAGTATATATGTCGCTTAGGAGGTTCCAGTTACTCAGCAGTTGCTTACATTTTAATAATTCCAAGAACTTGAGAAACCTCAACCTGGATCCCTCACAGGCAAAGCTTTTTAAAGTAGAGCCTGTGATTAGCCATTTGAATTCCAAGTTCACGGAGTTATATATAATGAAGCAGAACATGGCTTTAGATGAATCGCTGCTGCAGTGGAAATGTTGGTTGAACATCAACCAATTTATTCCAAACAAAGCTGCAGCAGTGGGCATAAAAACTTACGAAATATTTCTTAACCTTATCAAAGGTTTAGAACACAAAGGGTACACGTTGTGGATGGATAACTTCTACAATTCCCCAGCACTTGCCCGAAAACTCAAGTCGATTGGGTTTGATTGTGTCGGTACGTTGCGGACAAACCGCAAGTATGTCCCGACGGAACTGACCAATCTGAAAAAATCTCAGATGAAGCCCGGGCAAGTAGTAGGCTATACCAGTGGAGATGTCGATTGCATAATATGGAGAGATCAGAACCGCGTAGCGACGATCTCAACGTACCATGGCAATGCCGTCTCCACTAAAAACGGAGTAACAAAACTTATTTTGATACGTGACTACAACATCTGCATGGGTGGCGTGGATAAAAAAGATCAGATGTTGGCTGCATTCCCAATTGAACGCAAGAGGACACAAATTTGGTACGAGAAATTGTTTAAAAGGTTACTTAATGTTTCTGTGTTGAATGCCTATATCATACATAAACAGACTGCCACGGAGGTCTTGGACCACAGGGGCTTTAGAAAAAACCTGGTAGAATCCCTTTTGCGTAGGCATTCAATGAAAATGTAG
Protein
MTRSSLSYKCDCIYRFNLIAIEDLPAGEPIEQALTERSKRSWNPRNYEPDGEDIPVILSSPAIPPSPAMYVECGGSGTSTPSSRYNYRFLYMAAVVLCRHINVCVDRFAQTKNMILRTEILFHRYVDRYETEICYLLHYGSDDDDSDDESQNDNDEIVPMIIPRAAGILMDTPDDEEIEIDTAAASKPLVTSSSPSIISAPTSPARDTGMKPSHFFEFDWGTFPDSPIPPTERRESFKENSGPTVTEYMEFIVQETNRYAQQLAAEMLDGGELQASSRITEWKETNVDELLVFFGILLAMGIVIKNRVEEYWNTEQNIFSTPGFKVYMSLRRFQLLSSCLHFNNSKNLRNLNLDPSQAKLFKVEPVISHLNSKFTELYIMKQNMALDESLLQWKCWLNINQFIPNKAAAVGIKTYEIFLNLIKGLEHKGYTLWMDNFYNSPALARKLKSIGFDCVGTLRTNRKYVPTELTNLKKSQMKPGQVVGYTSGDVDCIIWRDQNRVATISTYHGNAVSTKNGVTKLILIRDYNICMGGVDKKDQMLAAFPIERKRTQIWYEKLFKRLLNVSVLNAYIIHKQTATEVLDHRGFRKNLVESLLRRHSMKM
Summary
Uniprot
EMBL
Proteomes
Pfam
PF13843 DDE_Tnp_1_7
Interpro
IPR029526
PGBD
ProteinModelPortal
Ontologies
GO
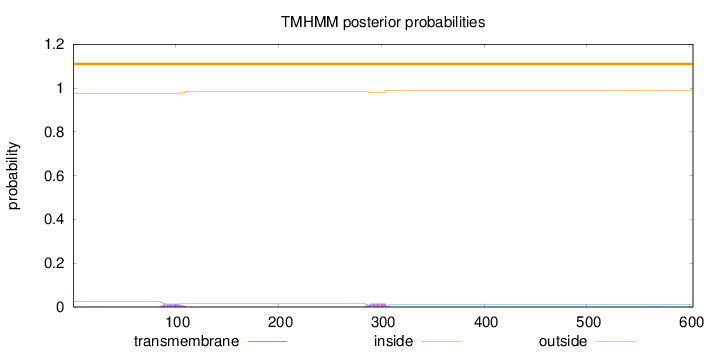
Topology
Length:
603
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.544769999999999
Exp number, first 60 AAs:
0.00321
Total prob of N-in:
0.02595
outside
1 - 603
Population Genetic Test Statistics
Pi
3.791157
Theta
4.668888
Tajima's D
-0.78498
CLR
0.133032
CSRT
0.182840857957102
Interpretation
Uncertain
