Gene
KWMTBOMO00445
Pre Gene Modal
BGIBMGA008616
Annotation
putative_DNA_helicase_[Cotesia_vestalis_bracovirus]
Full name
ATP-dependent DNA helicase
Location in the cell
Nuclear Reliability : 2.147
Sequence
CDS
ATGGGCAACATGGAAGAACAACTTGATCGACGCCTAGAGATCAATGCAGGAATGAAGCGAGCAATTCTTCAAGACTTGCAGTGTCTGCTTCATGAACATCATGCGTTGGTCAGGTTGTTTAAGAGTGCTTTAGAGCGCATGCCAAGTGATGACTATAAAGTTGTTATCAAAGCAGATAAACGACCATCTGGAACACACGAAAGGACATTTAATGCTCTAACAGTAGACGAAGTTGCCATCCTGATTGTTGGTGAACAATTGGAAAAACGCGATATTGTGCTTACACGTCGCGATACTGGGCAACTGCAACAAATATCTGAAACTCATCGATCATATGACACATTGCAATACCCGCTGATGTTTTGGCAAGGAGAAGATGGATATTATTTTAATATTAAAATGAAAAATCCATTGAATGGTGAAGAAACTACAAAGAAAGTTAGCTCAATGAATTATTACGCATATCGTTTGATGATTCGTCAAAATGCTGACAACTATTTGCTGCGGTTTCGTCGATTGTTTCAGCAGTATTGCGTTGACATGTATGTAAAAATAGAAACGGAACGTTTAACATTCATTAGGTTGAACCAAGCCAAACTGCGTTCTGAGGAGTACATCCATTTACGTGATGCAGTTAGTACTGAAGGAAATGCAGCTAATATTGGTCGATTAACTATTCTGCCGGCGACATACATTGGTAGTCCACGTCATATGCATGAATATGCACAAGATGCAATGACATATGTTCGTCATTACGGTCGGCCTGATCTCTTTATTACTTTTACCTGTAATCCAAAATGGATAGAAATTACTCAATTGCTGCTTCCCGGACAAACATCAAGTGATAGACACGACATCACAGCACGTATATTCAGGCAAAAAATTCGGTCCCTGATGAACTTTATTGTTAAACAACGCGTCTTTGGAGATACTCGATGCTGGATGTATTCAATCGAATGGCAAAAGCGAGGCCTGCCGCACGCACACATTCTTATTTGGTTAGTGGAAAGAATTCAGCCTGACCAAATAGATGATATCATATGTGCTGAGATTCCTGATTATGAAGTCGATCCAGACCTACATGATGTTGTTACTACTAATATGATTCATGGACCGTGTGGTGCCATCAATCCCCAATCACCTTGCATGGTCGATGGAAAGTGCTCTAAACGATACCCACGGAAATTAACGGCGGAGACTGTCACTGGCAATGATGGTTATCCGCTGTTAGGAATGCCAGCACCATATCGTGAAATGAATGACGCATTTAATCGAGAATTGGAAAGGGAACGTGAATATGATCACCAGGAATTAGATTTAGTAGTTCAAACGAATGTACCCCTGTTGAATTACCAACAAAAGGAAGTTTATGATACTTTAATGAAGGCAATCGATGATGAAAATGGTGGTTTATATTTTCTAGATGCCCCTGGTGGAACTGGCAAGACATTCCTCATGTCATTAGTTTTAGCAACTGTTCGGGCGAGATTTAACATAGCGGTTGCAGTTGCTTCTTCTGGAATAGCAGCCACATTGTTAGAAGGATGCCGTACGGCTCATTCAGCATTCAAATTACCGTTAAATCTTCAAACTATTGAAGAACCAACGTGTAATATTGCAAAACACTCAGCAATGGCCAAAGTTTTAGCGAGGACCTAA
Protein
MGNMEEQLDRRLEINAGMKRAILQDLQCLLHEHHALVRLFKSALERMPSDDYKVVIKADKRPSGTHERTFNALTVDEVAILIVGEQLEKRDIVLTRRDTGQLQQISETHRSYDTLQYPLMFWQGEDGYYFNIKMKNPLNGEETTKKVSSMNYYAYRLMIRQNADNYLLRFRRLFQQYCVDMYVKIETERLTFIRLNQAKLRSEEYIHLRDAVSTEGNAANIGRLTILPATYIGSPRHMHEYAQDAMTYVRHYGRPDLFITFTCNPKWIEITQLLLPGQTSSDRHDITARIFRQKIRSLMNFIVKQRVFGDTRCWMYSIEWQKRGLPHAHILIWLVERIQPDQIDDIICAEIPDYEVDPDLHDVVTTNMIHGPCGAINPQSPCMVDGKCSKRYPRKLTAETVTGNDGYPLLGMPAPYREMNDAFNRELEREREYDHQELDLVVQTNVPLLNYQQKEVYDTLMKAIDDENGGLYFLDAPGGTGKTFLMSLVLATVRARFNIAVAVASSGIAATLLEGCRTAHSAFKLPLNLQTIEEPTCNIAKHSAMAKVLART
Summary
Catalytic Activity
ATP + H2O = ADP + H(+) + phosphate
Cofactor
Mg(2+)
Similarity
Belongs to the helicase family.
Uniprot
F4ZH17
J9L7H1
E2AZ86
A0A0B7BHJ6
A0A0B7BEB0
A0A0C2GYY1
+ More
A0A0D6L4E4 A0A0C2CWG5 A0A016W7D9 A0A368GX53 A0A0C2G6C2 A0A0D6L8U7 A0A016SC70 A0A368GH99 A0A0C2CV48 A0A016VYY5 A0A182HPH8 A0A0D6L9G2 A0A0C2FKB3 A0A0C2CCC7 A0A267E3S5 A0A267FMY4 A0A267F5E2 A0A267F2R0 A0A182NCC1 A0A267GWL2 A0A267GEW9 A0A267GN76 J9KAP3 A0A267F4D8 A0A0C2CBQ7 A0A2H1W8I9 A0A1B6GN63 A0A1S3DI71 A0A2H1X3U0 J9LB24 J9LDS3 A0A0D6LBD4 A0A016SFH2 X1X109 J9LR77 A0A267H914 A0A267DEU7 A0A267F6F6 A0A267EIS0 A0A267GJF9 A0A267E9Q5 A0A267DB74 A0A267EKI4 A0A267ESS5 A0A267H554 A0A267G464 A0A267DG55 A0A267E2L2 A0A267FSI1 A0A267GX15 A0A267G7D2 A0A267GFW5 A0A267E6L9 A0A267GEV7 A0A267GJ34 A0A267G2P9
A0A0D6L4E4 A0A0C2CWG5 A0A016W7D9 A0A368GX53 A0A0C2G6C2 A0A0D6L8U7 A0A016SC70 A0A368GH99 A0A0C2CV48 A0A016VYY5 A0A182HPH8 A0A0D6L9G2 A0A0C2FKB3 A0A0C2CCC7 A0A267E3S5 A0A267FMY4 A0A267F5E2 A0A267F2R0 A0A182NCC1 A0A267GWL2 A0A267GEW9 A0A267GN76 J9KAP3 A0A267F4D8 A0A0C2CBQ7 A0A2H1W8I9 A0A1B6GN63 A0A1S3DI71 A0A2H1X3U0 J9LB24 J9LDS3 A0A0D6LBD4 A0A016SFH2 X1X109 J9LR77 A0A267H914 A0A267DEU7 A0A267F6F6 A0A267EIS0 A0A267GJF9 A0A267E9Q5 A0A267DB74 A0A267EKI4 A0A267ESS5 A0A267H554 A0A267G464 A0A267DG55 A0A267E2L2 A0A267FSI1 A0A267GX15 A0A267G7D2 A0A267GFW5 A0A267E6L9 A0A267GEV7 A0A267GJ34 A0A267G2P9
EC Number
3.6.4.12
EMBL
HQ009558
AEE09607.1
ABLF02021131
ABLF02042289
GL444143
EFN61253.1
+ More
HACG01044765 CEK91630.1 HACG01044764 CEK91629.1 KN727000 KIH66715.1 KE126587 EPB66060.1 KN739487 KIH54222.1 JARK01000714 EYC35207.1 JOJR01000041 RCN48933.1 KN743176 KIH52626.1 KE125664 EPB67478.1 JARK01001590 EYB87952.1 JOJR01000205 RCN42227.1 KN740537 KIH53717.1 JARK01001339 EYC32590.1 APCN01007923 KE125416 EPB68565.1 KN768033 KIH47189.1 KN767628 KIH47497.1 NIVC01002773 PAA55312.1 NIVC01000900 PAA75136.1 NIVC01001359 PAA68995.1 NIVC01001486 PAA67379.1 NIVC01000111 PAA90396.1 NIVC01000366 PAA84583.1 NIVC01000229 PAA87485.1 ABLF02006716 ABLF02036458 NIVC01001445 PAA67902.1 KN740416 KIH53763.1 ODYU01007016 SOQ49380.1 GECZ01005881 JAS63888.1 ODYU01013296 SOQ60003.1 ABLF02018127 ABLF02006224 ABLF02066369 KE125418 EPB68548.1 JARK01001572 EYB89086.1 ABLF02020814 ABLF02049665 ABLF02011507 ABLF02067513 NIVC01000001 PAA94775.1 NIVC01004365 PAA47686.1 NIVC01001383 PAA68602.1 NIVC01002034 PAA61410.1 NIVC01000291 PAA86185.1 NIVC01002407 PAA58126.1 NIVC01004811 PAA46541.1 NIVC01002028 PAA61494.1 NIVC01002018 NIVC01001743 NIVC01000051 PAA61561.1 PAA64516.1 PAA92464.1 NIVC01000042 PAA92669.1 NIVC01000566 PAA80831.1 NIVC01004209 PAA48195.1 NIVC01002707 PAA55791.1 NIVC01000853 PAA75959.1 NIVC01000136 PAA89829.1 NIVC01000506 PAA81945.1 NIVC01000353 PAA84930.1 NIVC01002606 PAA56554.1 PAA84578.1 NIVC01000296 PAA86068.1 NIVC01000587 PAA80345.1
HACG01044765 CEK91630.1 HACG01044764 CEK91629.1 KN727000 KIH66715.1 KE126587 EPB66060.1 KN739487 KIH54222.1 JARK01000714 EYC35207.1 JOJR01000041 RCN48933.1 KN743176 KIH52626.1 KE125664 EPB67478.1 JARK01001590 EYB87952.1 JOJR01000205 RCN42227.1 KN740537 KIH53717.1 JARK01001339 EYC32590.1 APCN01007923 KE125416 EPB68565.1 KN768033 KIH47189.1 KN767628 KIH47497.1 NIVC01002773 PAA55312.1 NIVC01000900 PAA75136.1 NIVC01001359 PAA68995.1 NIVC01001486 PAA67379.1 NIVC01000111 PAA90396.1 NIVC01000366 PAA84583.1 NIVC01000229 PAA87485.1 ABLF02006716 ABLF02036458 NIVC01001445 PAA67902.1 KN740416 KIH53763.1 ODYU01007016 SOQ49380.1 GECZ01005881 JAS63888.1 ODYU01013296 SOQ60003.1 ABLF02018127 ABLF02006224 ABLF02066369 KE125418 EPB68548.1 JARK01001572 EYB89086.1 ABLF02020814 ABLF02049665 ABLF02011507 ABLF02067513 NIVC01000001 PAA94775.1 NIVC01004365 PAA47686.1 NIVC01001383 PAA68602.1 NIVC01002034 PAA61410.1 NIVC01000291 PAA86185.1 NIVC01002407 PAA58126.1 NIVC01004811 PAA46541.1 NIVC01002028 PAA61494.1 NIVC01002018 NIVC01001743 NIVC01000051 PAA61561.1 PAA64516.1 PAA92464.1 NIVC01000042 PAA92669.1 NIVC01000566 PAA80831.1 NIVC01004209 PAA48195.1 NIVC01002707 PAA55791.1 NIVC01000853 PAA75959.1 NIVC01000136 PAA89829.1 NIVC01000506 PAA81945.1 NIVC01000353 PAA84930.1 NIVC01002606 PAA56554.1 PAA84578.1 NIVC01000296 PAA86068.1 NIVC01000587 PAA80345.1
Proteomes
Interpro
ProteinModelPortal
F4ZH17
J9L7H1
E2AZ86
A0A0B7BHJ6
A0A0B7BEB0
A0A0C2GYY1
+ More
A0A0D6L4E4 A0A0C2CWG5 A0A016W7D9 A0A368GX53 A0A0C2G6C2 A0A0D6L8U7 A0A016SC70 A0A368GH99 A0A0C2CV48 A0A016VYY5 A0A182HPH8 A0A0D6L9G2 A0A0C2FKB3 A0A0C2CCC7 A0A267E3S5 A0A267FMY4 A0A267F5E2 A0A267F2R0 A0A182NCC1 A0A267GWL2 A0A267GEW9 A0A267GN76 J9KAP3 A0A267F4D8 A0A0C2CBQ7 A0A2H1W8I9 A0A1B6GN63 A0A1S3DI71 A0A2H1X3U0 J9LB24 J9LDS3 A0A0D6LBD4 A0A016SFH2 X1X109 J9LR77 A0A267H914 A0A267DEU7 A0A267F6F6 A0A267EIS0 A0A267GJF9 A0A267E9Q5 A0A267DB74 A0A267EKI4 A0A267ESS5 A0A267H554 A0A267G464 A0A267DG55 A0A267E2L2 A0A267FSI1 A0A267GX15 A0A267G7D2 A0A267GFW5 A0A267E6L9 A0A267GEV7 A0A267GJ34 A0A267G2P9
A0A0D6L4E4 A0A0C2CWG5 A0A016W7D9 A0A368GX53 A0A0C2G6C2 A0A0D6L8U7 A0A016SC70 A0A368GH99 A0A0C2CV48 A0A016VYY5 A0A182HPH8 A0A0D6L9G2 A0A0C2FKB3 A0A0C2CCC7 A0A267E3S5 A0A267FMY4 A0A267F5E2 A0A267F2R0 A0A182NCC1 A0A267GWL2 A0A267GEW9 A0A267GN76 J9KAP3 A0A267F4D8 A0A0C2CBQ7 A0A2H1W8I9 A0A1B6GN63 A0A1S3DI71 A0A2H1X3U0 J9LB24 J9LDS3 A0A0D6LBD4 A0A016SFH2 X1X109 J9LR77 A0A267H914 A0A267DEU7 A0A267F6F6 A0A267EIS0 A0A267GJF9 A0A267E9Q5 A0A267DB74 A0A267EKI4 A0A267ESS5 A0A267H554 A0A267G464 A0A267DG55 A0A267E2L2 A0A267FSI1 A0A267GX15 A0A267G7D2 A0A267GFW5 A0A267E6L9 A0A267GEV7 A0A267GJ34 A0A267G2P9
Ontologies
GO
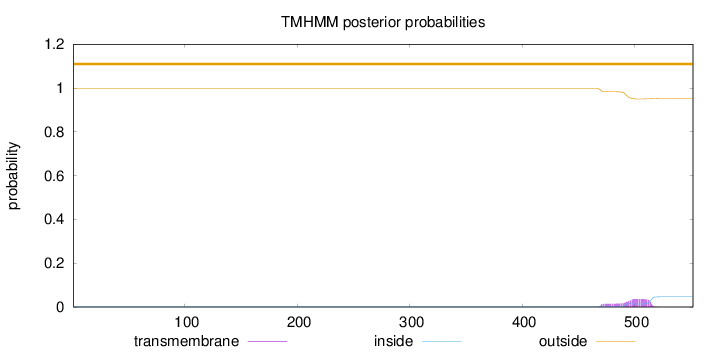
Topology
Length:
552
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
1.09772
Exp number, first 60 AAs:
0.00017
Total prob of N-in:
0.00235
outside
1 - 552
Population Genetic Test Statistics
Pi
2.790083
Theta
2.430597
Tajima's D
-1.300247
CLR
0.005568
CSRT
0.0782960851957402
Interpretation
Uncertain
