Gene
KWMTBOMO00443
Pre Gene Modal
BGIBMGA000480
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_protein_scabrous_[Bombyx_mori]
Full name
Protein scabrous
Location in the cell
Nuclear Reliability : 2.862
Sequence
CDS
ATGTTGTGCCCGAAAATTTGTGGACTCTTGACTTTATTTCTTGTTTTCACCGTGATCAAGGCCGAGTTGAGGTTCAAGGAAGAGCTGCAGGCCGTCGCTGAACAAGTCGAGATCATGAAAACTTTGCACCTTACTGACATTACTCGTCTGAAAAGCGAGATTGAGGAGCTCAAACACAAACGTTCTACGCCAACCAACAATTACGAGAGAAATGAACAGGCAACACTCCAATGGGCCAAGACCTCCATGAAAGAGCTGAAGATGGAGATCAGGGAAATCGGTGAAAGCGTCAACAACTCAGCCCTCCTCCGTCAACTGCACATCATACGTAATGAGCTAAAGCAAGCCCTTACCGAGACCTCGGACTTAGCGCAACTTGCTCGCACTCAAGAAGCCAGGCTAGACAAGATGGACAATGAAGTTGGCAGGCTCAAATACGAAGGACAACAGATGAGGAGTACTGTGGCTGAATTACGAACCCACGTCAGCAAGCTGGTCAAAGAGATGAAAGCTAAACAAGTGGCGGAGATGAACGGTGACACCTACAATGATGTAATTGAGCCCCACGAGAACCGAGCCGGTGATCTTAAGCCCGAGCGCAACCATAAGTACCGCCACAACAAGATGGTGCACGCTCAGATTTCACGACTTGCACGCAGCCAGGAGCAGCTTGACGAATACCAACAGAGCCTACAGACCCAGATCCTTGATGTGCAGCGCCGGCTCGACCGCTTCGAAGAACCGAACTGGAACCTACTTTCAGCCAAGGTCGATTTCCTCGAAGTTGAATCGAAAATACTTCGGAACGAACTGCAGAACACCACACAGAGAGTGTCCGATTTCGATAAAGTGCACGCTTCCATTCTCGAGCTGCGTGAAGACATCGAAGGGCTCGAGAACAAAGTCGACAAAACTGTACCGGATTTCAGAAAGGAAATATCGAAATTGGACGTCAGCTTTGCCCAGCTGAATGCTCAGTCTTCGTACCTCAAGGAGGATCAGGAGAATATACGTCAATCAGTAAAAGCTATAGCAGTCAGTGTCAGCAACGCCATCGACCGCGCCGAGATGGACCGCCTGATGGTTAAGAAGATCAACGACTCCGTGTTCGACTTGGAAGCCCGGTCCAAGCAGCATTACTACAGACTCAACGATCACATTCTCAAGAGCGAGGCCAGTGTTCCAGACGCTAATTTGACTGAAACTATACCCCTTCCGGAGCTGATGGGCGAGGTCAAGGAACTGGAGCCGGTCCAACGTGAGTACGAGGATCTAGTCAACCAGCTACCTCGTGACTGCTCCAGCATCAAGGGGCCGGCCGGGACGTACCTTATCCACCCTGGGCACGCTCCCGTCGACACCTGGTGCGTCAACGGAAACACTCTATTGCAAAGACGCTATAATGGCTCGATCGAATTCAATCGTAAATTTAGCGAGTACGTTCATGGTTTCGGCGACCCCACTTCGGAATACTGGTTGGGCTTGGAATCAATGCACCAGATCACTTCCGATAACTGCTCTTCGATGAGGATCGATATGACCGATATATACGGCGGCAGCTGGTATGCCGAATACGATCACTTCTCGGTCAGCAACGCCGACACCGGATTCATTCTGAACGTGAGCGGATTTAGAGGAAATGCCAGCGATGCTTTTGAGTATCAAAATCATATGGAGTTCTCGACTATCGACCGCGACCGCGACATCTCTAACACTCACTGCGCCGGCAACTACGAGGGCGGTTGGTGGTTCTCTCACTGCCAGCATGTGAACATCAACGGCAAGTACACACTCGGCCTGACCTGGTTCGACTCCGCCAGGAACGAGTGGATCGCCGTAGCCACTAGCGAGATGCGCGTGTTCCGCAGAGCCGGCTGCTTCTAA
Protein
MLCPKICGLLTLFLVFTVIKAELRFKEELQAVAEQVEIMKTLHLTDITRLKSEIEELKHKRSTPTNNYERNEQATLQWAKTSMKELKMEIREIGESVNNSALLRQLHIIRNELKQALTETSDLAQLARTQEARLDKMDNEVGRLKYEGQQMRSTVAELRTHVSKLVKEMKAKQVAEMNGDTYNDVIEPHENRAGDLKPERNHKYRHNKMVHAQISRLARSQEQLDEYQQSLQTQILDVQRRLDRFEEPNWNLLSAKVDFLEVESKILRNELQNTTQRVSDFDKVHASILELREDIEGLENKVDKTVPDFRKEISKLDVSFAQLNAQSSYLKEDQENIRQSVKAIAVSVSNAIDRAEMDRLMVKKINDSVFDLEARSKQHYYRLNDHILKSEASVPDANLTETIPLPELMGEVKELEPVQREYEDLVNQLPRDCSSIKGPAGTYLIHPGHAPVDTWCVNGNTLLQRRYNGSIEFNRKFSEYVHGFGDPTSEYWLGLESMHQITSDNCSSMRIDMTDIYGGSWYAEYDHFSVSNADTGFILNVSGFRGNASDAFEYQNHMEFSTIDRDRDISNTHCAGNYEGGWWFSHCQHVNINGKYTLGLTWFDSARNEWIAVATSEMRVFRRAGCF
Summary
Description
Involved in regulation of neurogenesis. May encode a lateral inhibitor of R8 differentiation. In conjunction with Gp150, promotes Notch activation in response to Delta by regulating acquisition of insensitivity to Delta in a subset of cells.
Keywords
Complete proteome
Developmental protein
Differentiation
Disulfide bond
Endosome
Glycoprotein
Neurogenesis
Notch signaling pathway
Reference proteome
Signal
Feature
chain Protein scabrous
Uniprot
A0A2A4JSI9
A0A2W1C0K7
A0A212F9J7
A0A194PGS3
A0A194QLF3
A0A2H1WLZ3
+ More
A0A2A4JRC5 A0A0L7KVL0 H9ITA0 A0A1W4XNJ2 A0A1Y1LMR7 D6WEF6 A0A2J7QQM2 N6TWG4 A0A1B6D1F2 A0A067RJR4 Q16TT3 A0A1S4FPK1 A0A182QUW6 A0A182Y288 A0A1Y9IUZ9 Q5TWN1 A0A1S4GTY1 B0X0D0 A0A182JAF2 A0A084WRL1 A0A3F2YXN1 A0A1B0CV60 W5JUA4 B4LK98 A0A336MJT6 B4J6U0 B3MD35 Q28XV4 B4GH40 B4MX41 A0A1B0FF50 A0A1A9Z5M0 A0A1A9UI60 A0A1B0BGC2 A0A1A9XG10 A0A3B0J2W6 A0A1I8PVJ1 A0A1W4VDB7 B4KNF8 P21520 A0A0J9U085 B4P4Y6 B3NRV2 B4QDH3 B4HPP5 A0A0M4EHV8 A0A0K8VMS8 A0A0L0BMC4 A0A146LY72 A0A2P8ZDB0 A0A2H8TGX7 A0A1J1JBR2 J9JKY9 A0A3Q0ILC5 A0A0C9RT39 A0A026WSH0 A0A3L8D9F8 A0A195CNV5 E2AZF8 T1HD12 E9IB88 A0A158NI54 A0A195B674 A0A195EGQ9 A0A151JXD2 A0A0J7LAG9 A0A0C9R307 F4WGB4 A0A0L7QSL1 A0A164YA83 A0A0P5LP28 A0A0P6AY65 A0A0M9A6Z2
A0A2A4JRC5 A0A0L7KVL0 H9ITA0 A0A1W4XNJ2 A0A1Y1LMR7 D6WEF6 A0A2J7QQM2 N6TWG4 A0A1B6D1F2 A0A067RJR4 Q16TT3 A0A1S4FPK1 A0A182QUW6 A0A182Y288 A0A1Y9IUZ9 Q5TWN1 A0A1S4GTY1 B0X0D0 A0A182JAF2 A0A084WRL1 A0A3F2YXN1 A0A1B0CV60 W5JUA4 B4LK98 A0A336MJT6 B4J6U0 B3MD35 Q28XV4 B4GH40 B4MX41 A0A1B0FF50 A0A1A9Z5M0 A0A1A9UI60 A0A1B0BGC2 A0A1A9XG10 A0A3B0J2W6 A0A1I8PVJ1 A0A1W4VDB7 B4KNF8 P21520 A0A0J9U085 B4P4Y6 B3NRV2 B4QDH3 B4HPP5 A0A0M4EHV8 A0A0K8VMS8 A0A0L0BMC4 A0A146LY72 A0A2P8ZDB0 A0A2H8TGX7 A0A1J1JBR2 J9JKY9 A0A3Q0ILC5 A0A0C9RT39 A0A026WSH0 A0A3L8D9F8 A0A195CNV5 E2AZF8 T1HD12 E9IB88 A0A158NI54 A0A195B674 A0A195EGQ9 A0A151JXD2 A0A0J7LAG9 A0A0C9R307 F4WGB4 A0A0L7QSL1 A0A164YA83 A0A0P5LP28 A0A0P6AY65 A0A0M9A6Z2
Pubmed
28756777
22118469
26354079
26227816
19121390
28004739
+ More
18362917 19820115 23537049 24845553 17510324 25244985 12364791 24438588 20920257 23761445 17994087 15632085 2175046 10731132 12537572 12537569 12756167 22936249 17550304 26108605 26823975 29403074 24508170 30249741 20798317 21282665 21347285 21719571
18362917 19820115 23537049 24845553 17510324 25244985 12364791 24438588 20920257 23761445 17994087 15632085 2175046 10731132 12537572 12537569 12756167 22936249 17550304 26108605 26823975 29403074 24508170 30249741 20798317 21282665 21347285 21719571
EMBL
NWSH01000761
PCG74370.1
KZ149901
PZC78496.1
AGBW02009564
OWR50412.1
+ More
KQ459605 KPI91904.1 KQ461196 KPJ06393.1 ODYU01009479 SOQ53952.1 PCG74369.1 JTDY01005153 KOB67312.1 BABH01007345 GEZM01053187 JAV74138.1 KQ971318 EFA01305.1 NEVH01012087 PNF30891.1 APGK01049655 APGK01049656 KB741156 ENN73590.1 GEDC01017790 JAS19508.1 KK852639 KDR19650.1 CH477640 EAT37938.1 AXCN02000438 AAAB01008807 EAL41889.3 DS232233 EDS38091.1 ATLV01026157 ATLV01026158 ATLV01026159 ATLV01026160 ATLV01026161 KE525407 KFB52855.1 AJWK01030133 AJWK01030134 AJWK01030135 AJWK01030136 ADMH02000472 ETN66339.1 CH940648 EDW61689.1 UFQT01001464 SSX30674.1 CH916367 EDW02021.1 CH902619 EDV36350.2 CM000071 EAL26212.2 CH479183 EDW35810.1 CH963857 EDW76680.1 CCAG010011452 JXJN01013822 OUUW01000001 SPP75607.1 CH933808 EDW08917.1 M60065 AE013599 AY129456 CM002911 KMY93340.1 CM000158 EDW90707.1 CH954179 EDV56254.1 CM000362 EDX06838.1 CH480816 EDW47629.1 CP012524 ALC40567.1 GDHF01012118 JAI40196.1 JRES01001648 KNC21196.1 GDHC01006116 JAQ12513.1 PYGN01000092 PSN54485.1 GFXV01001549 MBW13354.1 CVRI01000075 CRL08513.1 ABLF02029629 GBYB01010671 JAG80438.1 KK107111 EZA58995.1 QOIP01000011 RLU16782.1 KQ977481 KYN02418.1 GL444207 EFN61197.1 ACPB03009095 GL762111 EFZ22173.1 ADTU01016299 KQ976579 KYM80018.1 KQ978957 KYN27069.1 KQ981578 KYN39762.1 LBMM01000112 KMR04882.1 GBYB01002315 JAG72082.1 GL888128 EGI66881.1 KQ414758 KOC61456.1 LRGB01000930 KZS15043.1 GDIQ01170566 JAK81159.1 GDIP01029292 JAM74423.1 KQ435724 KOX78497.1
KQ459605 KPI91904.1 KQ461196 KPJ06393.1 ODYU01009479 SOQ53952.1 PCG74369.1 JTDY01005153 KOB67312.1 BABH01007345 GEZM01053187 JAV74138.1 KQ971318 EFA01305.1 NEVH01012087 PNF30891.1 APGK01049655 APGK01049656 KB741156 ENN73590.1 GEDC01017790 JAS19508.1 KK852639 KDR19650.1 CH477640 EAT37938.1 AXCN02000438 AAAB01008807 EAL41889.3 DS232233 EDS38091.1 ATLV01026157 ATLV01026158 ATLV01026159 ATLV01026160 ATLV01026161 KE525407 KFB52855.1 AJWK01030133 AJWK01030134 AJWK01030135 AJWK01030136 ADMH02000472 ETN66339.1 CH940648 EDW61689.1 UFQT01001464 SSX30674.1 CH916367 EDW02021.1 CH902619 EDV36350.2 CM000071 EAL26212.2 CH479183 EDW35810.1 CH963857 EDW76680.1 CCAG010011452 JXJN01013822 OUUW01000001 SPP75607.1 CH933808 EDW08917.1 M60065 AE013599 AY129456 CM002911 KMY93340.1 CM000158 EDW90707.1 CH954179 EDV56254.1 CM000362 EDX06838.1 CH480816 EDW47629.1 CP012524 ALC40567.1 GDHF01012118 JAI40196.1 JRES01001648 KNC21196.1 GDHC01006116 JAQ12513.1 PYGN01000092 PSN54485.1 GFXV01001549 MBW13354.1 CVRI01000075 CRL08513.1 ABLF02029629 GBYB01010671 JAG80438.1 KK107111 EZA58995.1 QOIP01000011 RLU16782.1 KQ977481 KYN02418.1 GL444207 EFN61197.1 ACPB03009095 GL762111 EFZ22173.1 ADTU01016299 KQ976579 KYM80018.1 KQ978957 KYN27069.1 KQ981578 KYN39762.1 LBMM01000112 KMR04882.1 GBYB01002315 JAG72082.1 GL888128 EGI66881.1 KQ414758 KOC61456.1 LRGB01000930 KZS15043.1 GDIQ01170566 JAK81159.1 GDIP01029292 JAM74423.1 KQ435724 KOX78497.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000053240
UP000037510
UP000005204
+ More
UP000192223 UP000007266 UP000235965 UP000019118 UP000027135 UP000008820 UP000075886 UP000076408 UP000075920 UP000007062 UP000002320 UP000075880 UP000030765 UP000075900 UP000092461 UP000000673 UP000008792 UP000001070 UP000007801 UP000001819 UP000008744 UP000007798 UP000092444 UP000092445 UP000078200 UP000092460 UP000092443 UP000268350 UP000095300 UP000192221 UP000009192 UP000000803 UP000002282 UP000008711 UP000000304 UP000001292 UP000092553 UP000037069 UP000245037 UP000183832 UP000007819 UP000079169 UP000053097 UP000279307 UP000078542 UP000000311 UP000015103 UP000005205 UP000078540 UP000078492 UP000078541 UP000036403 UP000007755 UP000053825 UP000076858 UP000053105
UP000192223 UP000007266 UP000235965 UP000019118 UP000027135 UP000008820 UP000075886 UP000076408 UP000075920 UP000007062 UP000002320 UP000075880 UP000030765 UP000075900 UP000092461 UP000000673 UP000008792 UP000001070 UP000007801 UP000001819 UP000008744 UP000007798 UP000092444 UP000092445 UP000078200 UP000092460 UP000092443 UP000268350 UP000095300 UP000192221 UP000009192 UP000000803 UP000002282 UP000008711 UP000000304 UP000001292 UP000092553 UP000037069 UP000245037 UP000183832 UP000007819 UP000079169 UP000053097 UP000279307 UP000078542 UP000000311 UP000015103 UP000005205 UP000078540 UP000078492 UP000078541 UP000036403 UP000007755 UP000053825 UP000076858 UP000053105
Pfam
PF00147 Fibrinogen_C
Interpro
SUPFAM
SSF56496
SSF56496
Gene 3D
CDD
ProteinModelPortal
A0A2A4JSI9
A0A2W1C0K7
A0A212F9J7
A0A194PGS3
A0A194QLF3
A0A2H1WLZ3
+ More
A0A2A4JRC5 A0A0L7KVL0 H9ITA0 A0A1W4XNJ2 A0A1Y1LMR7 D6WEF6 A0A2J7QQM2 N6TWG4 A0A1B6D1F2 A0A067RJR4 Q16TT3 A0A1S4FPK1 A0A182QUW6 A0A182Y288 A0A1Y9IUZ9 Q5TWN1 A0A1S4GTY1 B0X0D0 A0A182JAF2 A0A084WRL1 A0A3F2YXN1 A0A1B0CV60 W5JUA4 B4LK98 A0A336MJT6 B4J6U0 B3MD35 Q28XV4 B4GH40 B4MX41 A0A1B0FF50 A0A1A9Z5M0 A0A1A9UI60 A0A1B0BGC2 A0A1A9XG10 A0A3B0J2W6 A0A1I8PVJ1 A0A1W4VDB7 B4KNF8 P21520 A0A0J9U085 B4P4Y6 B3NRV2 B4QDH3 B4HPP5 A0A0M4EHV8 A0A0K8VMS8 A0A0L0BMC4 A0A146LY72 A0A2P8ZDB0 A0A2H8TGX7 A0A1J1JBR2 J9JKY9 A0A3Q0ILC5 A0A0C9RT39 A0A026WSH0 A0A3L8D9F8 A0A195CNV5 E2AZF8 T1HD12 E9IB88 A0A158NI54 A0A195B674 A0A195EGQ9 A0A151JXD2 A0A0J7LAG9 A0A0C9R307 F4WGB4 A0A0L7QSL1 A0A164YA83 A0A0P5LP28 A0A0P6AY65 A0A0M9A6Z2
A0A2A4JRC5 A0A0L7KVL0 H9ITA0 A0A1W4XNJ2 A0A1Y1LMR7 D6WEF6 A0A2J7QQM2 N6TWG4 A0A1B6D1F2 A0A067RJR4 Q16TT3 A0A1S4FPK1 A0A182QUW6 A0A182Y288 A0A1Y9IUZ9 Q5TWN1 A0A1S4GTY1 B0X0D0 A0A182JAF2 A0A084WRL1 A0A3F2YXN1 A0A1B0CV60 W5JUA4 B4LK98 A0A336MJT6 B4J6U0 B3MD35 Q28XV4 B4GH40 B4MX41 A0A1B0FF50 A0A1A9Z5M0 A0A1A9UI60 A0A1B0BGC2 A0A1A9XG10 A0A3B0J2W6 A0A1I8PVJ1 A0A1W4VDB7 B4KNF8 P21520 A0A0J9U085 B4P4Y6 B3NRV2 B4QDH3 B4HPP5 A0A0M4EHV8 A0A0K8VMS8 A0A0L0BMC4 A0A146LY72 A0A2P8ZDB0 A0A2H8TGX7 A0A1J1JBR2 J9JKY9 A0A3Q0ILC5 A0A0C9RT39 A0A026WSH0 A0A3L8D9F8 A0A195CNV5 E2AZF8 T1HD12 E9IB88 A0A158NI54 A0A195B674 A0A195EGQ9 A0A151JXD2 A0A0J7LAG9 A0A0C9R307 F4WGB4 A0A0L7QSL1 A0A164YA83 A0A0P5LP28 A0A0P6AY65 A0A0M9A6Z2
PDB
4M7H
E-value=1.59884e-28,
Score=316
Ontologies
GO
PANTHER
Topology
Subcellular location
Late endosome
SignalP
Position: 1 - 21,
Likelihood: 0.909303
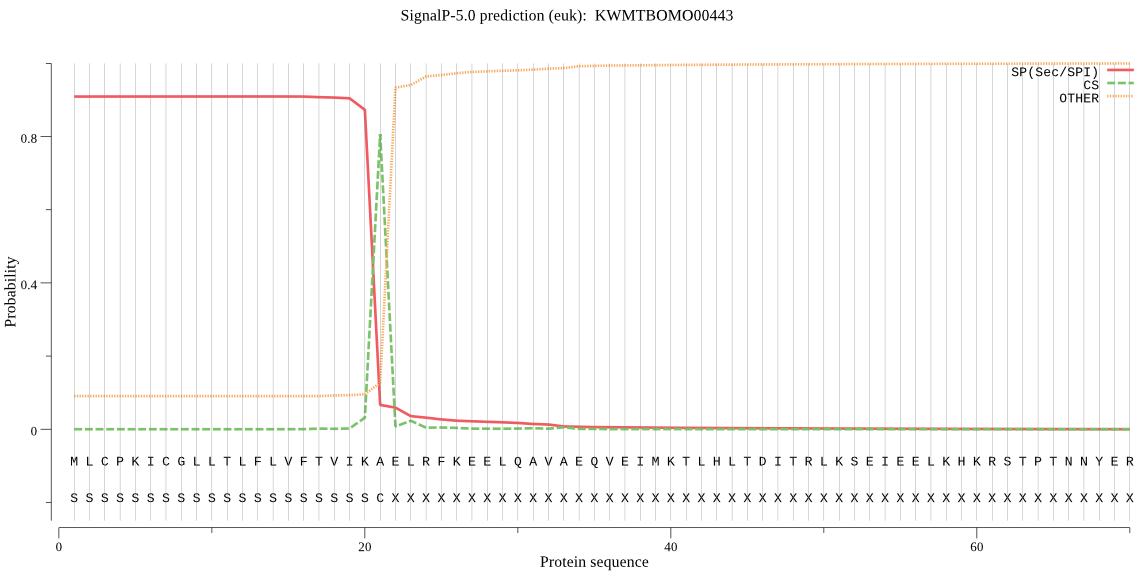
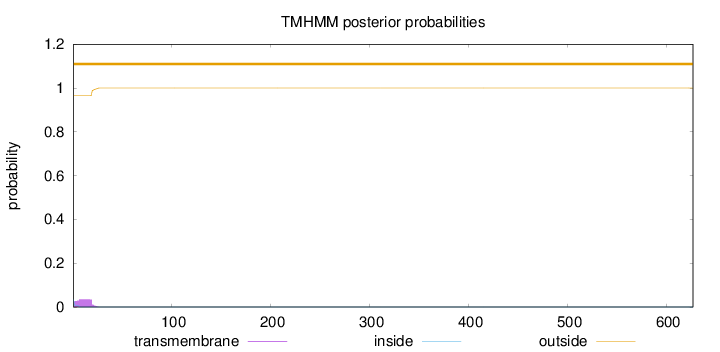
Length:
627
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.624249999999999
Exp number, first 60 AAs:
0.62406
Total prob of N-in:
0.03462
outside
1 - 627
Population Genetic Test Statistics
Pi
22.305147
Theta
18.469359
Tajima's D
0.492565
CLR
0.12446
CSRT
0.522673866306685
Interpretation
Uncertain
