Gene
KWMTBOMO00438 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000481
Annotation
Sulfhydryl_oxidase_1_[Papilio_machaon]
Full name
Sulfhydryl oxidase
Location in the cell
PlasmaMembrane Reliability : 1.945
Sequence
CDS
ATGAATTTCTGGTATACGTCTATTTTTGAAATATTTTTGATAGCTCTCGTTACGGGAGCAGTTGTGCCCACAACCGACGATGTAGACGAACAAGGATTGTACCGTAAGTCAGATCAAGTCGAAATCTTGACTAATAAAAATTTCGAAAAGAAATTATACGGCCAAAATAATGCTCTGTTGGTTCAGTTTTATAATAGTTACTGCGGTCATTGCCGAGCGTTTTCACCGAAATATAAAGCACTCGCATCTGACATAGCTCGTTGGAAAAAAGTTATTAAGTTAGCTGTTATTGATTGTTTTGTTGAAGAAAATAGTGAAATATGTAGACAATTTGAAGTAATGGCATACCCTTCCATACGATACTTCCACGAAAATTATATGAAGAGCAACTCAAATGTTGGAGAAAAAATGAACATTGCGGATACCGCAGAAAGACTTAAAAATCAATTGATTATAAAATTACAGGCAGAACAATCTATGGGACGTCTAATAATTGCTCCATCATTTAAGATTGAATCATATACATCCTATGCATCTGCATTACAAAGTGTACCTGGTGACATTGACTACATATTTTTAGTATTTGAAAATGATAATTCAACTATTGGATCACAAATAGCTTTAGATGTTGTTGACTATAAAAAAGTGAGAGTTAAGAGAGTATATGAAAATAGTGAATTAGCTCAAGTTGCTGGAGTTAAAAAAATTCCAAGTGTAGTTGCTCTAGAAAATAATTTACAAGCCACATTACTAACACCCAAACAGCCCACTGCACAAAATATATTAGAAGAGATAGATAGATTTTTGAAATCAAAAAATTATGTCTTTCCTCCGAAATATGCCAACATGAATGATATCAGTGATAGTTCTCAACAGAGAACATTTGTGCCAACCTCTGATGTTGCATACTATAATGATCTAGAAAAAACTTTAAAGGCAAGCTTGCACACTGAAATCACAAGGCACAAAACATTAACAGGTGAACCATTGCAGGCTTTATTAGACTACTTGGATGTTATTAAAACTTCGTTTCCATTTAGAGCCAATCTTGGAGAATATATAATGGACTTACATGCTACTTTAGCAGCAAGAAACAGCTGGACAGGTGGTGAAGTCTATGATTTAGTAAAAAGACTAGAAACTGCTCATGAACCAGTTTATATTACTAATTTAGAGTATGTTGGATGCAAAGGAAGTGAACCTAAATACAGAGGTTATACATGTGGACTGTGGACATTGTTCCATACTCTGACAGTCAACGCAGCACAAAAACCAGGTTCTGAAGGACCTAAAGTTCTTAAAGCAATGCATGGATATGTCAAAAACTTCTTTGGGTGTACTGAATGTGCTTCACATTTTCAAGCAATGGCAGCACGTAACAGAATATTTGATGTCAAAGAGAATAATAAAGCGGTTTTGTGGCTTTGGATTTCTCATAATGAAGTAAACCTTAGACTTGCGGGTGATGTCACAGAAGACCCTGAACATCCGAAAATTCAGTTTCCCAGTGCCACTAAATGCCCTGAATGCAGACTAGCTCAAGGAGCATGGAATCTAGTAGCAGTGTATGACTATCTTCAAAAGATGTATGGTGCCAACAACATCAAAGATGTAAGAAGAGCACAAGTGTCATCTGCTTCCAGCACCTTCTCCAATTTGGACATCGGGATGTTAAGTCTTTTGTACATATTATCTTTTATTTTCATTATTTTGGTTATAAAATACTTTTGGTCTAAACGTTTCTATAGAAAAAGACATTATAAAATGGGAATGGGCAAAGTATAG
Protein
MNFWYTSIFEIFLIALVTGAVVPTTDDVDEQGLYRKSDQVEILTNKNFEKKLYGQNNALLVQFYNSYCGHCRAFSPKYKALASDIARWKKVIKLAVIDCFVEENSEICRQFEVMAYPSIRYFHENYMKSNSNVGEKMNIADTAERLKNQLIIKLQAEQSMGRLIIAPSFKIESYTSYASALQSVPGDIDYIFLVFENDNSTIGSQIALDVVDYKKVRVKRVYENSELAQVAGVKKIPSVVALENNLQATLLTPKQPTAQNILEEIDRFLKSKNYVFPPKYANMNDISDSSQQRTFVPTSDVAYYNDLEKTLKASLHTEITRHKTLTGEPLQALLDYLDVIKTSFPFRANLGEYIMDLHATLAARNSWTGGEVYDLVKRLETAHEPVYITNLEYVGCKGSEPKYRGYTCGLWTLFHTLTVNAAQKPGSEGPKVLKAMHGYVKNFFGCTECASHFQAMAARNRIFDVKENNKAVLWLWISHNEVNLRLAGDVTEDPEHPKIQFPSATKCPECRLAQGAWNLVAVYDYLQKMYGANNIKDVRRAQVSSASSTFSNLDIGMLSLLYILSFIFIILVIKYFWSKRFYRKRHYKMGMGKV
Summary
Catalytic Activity
O2 + 2 R'C(R)SH = H2O2 + R'C(R)S-S(R)CR'
Cofactor
FAD
Feature
chain Sulfhydryl oxidase
Uniprot
H9ITA1
A0A194QN37
A0A2W1BU08
A0A2A4JQP4
A0A2A4JQR5
A0A2A4JRH9
+ More
A0A194PF51 A0A220K8N7 A0A0L7L9F1 A0A2H1W0E0 A0A212F9J4 A0A1W4XTZ0 A0A1Y1LI55 A0A1Y1LCT5 N6TVR9 V5G6I6 U4USZ4 V9PB47 A0A0N1ISZ6 A0A139WK54 A0A195C0V8 A0A154PPC5 D6WHZ3 A0A026VU03 A0A0J7KVA4 A0A195EYN4 F4W5S4 E2AAN0 A0A151X2K2 K7J8R1 A0A158P2R4 E2BAZ4 A0A151I2S1 A0A195DFB6 A0A088A7I5 A0A2A3ED61 A0A0L7QNG1 A0A2J7PGP5 A0A2P8XEJ0 T1PA99 A0A1I8NB55 A0A067REY5 B3MGW1 E0VW74 A0A0L0BP21 A0A1A9VDD5 Q292N7 B4GD11 B4LM00 A0A336K3J4 A0A3B0IZY0 A0A1L8EFD2 A0A1L8EFK0 B4P4K4 A0A0M4EFT4 A0A1B0F9H4 A0A1I8QCB5 B4J5B5 B3NRN7 A0A1A9ZG99 A0A1A9X3I5 A0A034VIE8 A0A0K8V571 Q7JQR3 A0A1B6FCT0 A0A1B0BXH4 A0A1A9YJI3 A0A1B6KLZ5 B4QE06 B4HQ33 A0A1W4UNU0 A0A0C9PXU2 A0A0A1XLR9 B4NLD8 A0A0C9RHA5 A0A0Q9WVL8 A0A139WDJ7 W8C0Z9 A0A023EU40 A0A023EVQ9 A0A023EVD5 A0A182HDX8 A0A182GB20 A0A1S4FVG8 Q16N96 Q9VD61 A0A2S2NV03 B4PKY1 B4QZB9 J9JRQ3 A0A2R7WRG6 E9IPK0 B3P8P9 Q4FDV9 D6WHZ4 A0A2S2R6H8 A0A3Q0JDK6 B3LVJ9 B0UXN0 A0A091QLP6
A0A194PF51 A0A220K8N7 A0A0L7L9F1 A0A2H1W0E0 A0A212F9J4 A0A1W4XTZ0 A0A1Y1LI55 A0A1Y1LCT5 N6TVR9 V5G6I6 U4USZ4 V9PB47 A0A0N1ISZ6 A0A139WK54 A0A195C0V8 A0A154PPC5 D6WHZ3 A0A026VU03 A0A0J7KVA4 A0A195EYN4 F4W5S4 E2AAN0 A0A151X2K2 K7J8R1 A0A158P2R4 E2BAZ4 A0A151I2S1 A0A195DFB6 A0A088A7I5 A0A2A3ED61 A0A0L7QNG1 A0A2J7PGP5 A0A2P8XEJ0 T1PA99 A0A1I8NB55 A0A067REY5 B3MGW1 E0VW74 A0A0L0BP21 A0A1A9VDD5 Q292N7 B4GD11 B4LM00 A0A336K3J4 A0A3B0IZY0 A0A1L8EFD2 A0A1L8EFK0 B4P4K4 A0A0M4EFT4 A0A1B0F9H4 A0A1I8QCB5 B4J5B5 B3NRN7 A0A1A9ZG99 A0A1A9X3I5 A0A034VIE8 A0A0K8V571 Q7JQR3 A0A1B6FCT0 A0A1B0BXH4 A0A1A9YJI3 A0A1B6KLZ5 B4QE06 B4HQ33 A0A1W4UNU0 A0A0C9PXU2 A0A0A1XLR9 B4NLD8 A0A0C9RHA5 A0A0Q9WVL8 A0A139WDJ7 W8C0Z9 A0A023EU40 A0A023EVQ9 A0A023EVD5 A0A182HDX8 A0A182GB20 A0A1S4FVG8 Q16N96 Q9VD61 A0A2S2NV03 B4PKY1 B4QZB9 J9JRQ3 A0A2R7WRG6 E9IPK0 B3P8P9 Q4FDV9 D6WHZ4 A0A2S2R6H8 A0A3Q0JDK6 B3LVJ9 B0UXN0 A0A091QLP6
EC Number
1.8.3.2
Pubmed
19121390
26354079
28756777
28630352
26227816
22118469
+ More
28004739 23537049 18362917 19820115 24508170 21719571 20798317 20075255 21347285 29403074 25315136 24845553 17994087 20566863 26108605 15632085 17550304 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25830018 24495485 24945155 26483478 17510324 21282665 15944345 23594743
28004739 23537049 18362917 19820115 24508170 21719571 20798317 20075255 21347285 29403074 25315136 24845553 17994087 20566863 26108605 15632085 17550304 25348373 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 22936249 25830018 24495485 24945155 26483478 17510324 21282665 15944345 23594743
EMBL
BABH01007337
KQ461196
KPJ06390.1
KZ149901
PZC78499.1
NWSH01000761
+ More
PCG74375.1 PCG74377.1 PCG74376.1 KQ459605 KPI91907.1 MF319731 ASJ26445.1 JTDY01002142 KOB72015.1 ODYU01005567 SOQ46541.1 AGBW02009564 OWR50415.1 GEZM01061174 JAV70707.1 GEZM01061173 JAV70708.1 APGK01050031 APGK01050032 KB741165 ENN73390.1 GALX01002771 JAB65695.1 KB632352 ERL93296.1 KC282396 AGX25153.1 KQ436033 KOX67532.1 KQ971331 KYB28312.1 KQ978379 KYM94477.1 KQ434978 KZC13020.1 EEZ99710.2 KK107921 EZA47207.1 LBMM01002888 KMQ94231.1 KQ981905 KYN33333.1 GL887695 EGI70400.1 GL438137 EFN69480.1 KQ982576 KYQ54667.1 ADTU01007399 ADTU01007400 ADTU01007401 GL446901 EFN87085.1 KQ976518 KYM82197.1 KQ980903 KYN11595.1 KZ288279 PBC29685.1 KQ414851 KOC60182.1 NEVH01025174 PNF15507.1 PYGN01002482 PSN30415.1 KA645070 AFP59699.1 KK852680 KDR18624.1 CH902619 EDV37879.1 DS235817 EEB17630.1 JRES01001578 KNC21807.1 CM000071 EAL24825.1 CH479181 EDW31549.1 CH940648 EDW59920.1 UFQS01000067 UFQT01000067 SSW98991.1 SSX19373.1 OUUW01000001 SPP73895.1 GFDG01001403 JAV17396.1 GFDG01001404 JAV17395.1 CM000158 EDW90643.1 CP012524 ALC41135.1 CCAG010000868 CH916367 EDW01757.1 CH954179 EDV56189.1 GAKP01017045 JAC41907.1 GDHF01018240 GDHF01012411 GDHF01009786 JAI34074.1 JAI39903.1 JAI42528.1 AE013599 BT004854 AAF58400.1 AAO45210.1 GECZ01021764 JAS48005.1 JXJN01022230 GEBQ01027536 JAT12441.1 CM000362 CM002911 EDX06900.1 KMY93452.1 CH480816 EDW47696.1 GBYB01006313 JAG76080.1 GBXI01002759 JAD11533.1 CH964272 EDW84341.1 GBYB01006311 JAG76078.1 KRG00013.1 KQ971358 KYB26006.1 GAMC01011216 JAB95339.1 GAPW01000673 JAC12925.1 GAPW01000431 JAC13167.1 GAPW01000646 JAC12952.1 JXUM01034905 JXUM01034906 KQ561040 KXJ79927.1 JXUM01051977 JXUM01051978 KQ561713 KXJ77746.1 CH477833 EAT35810.1 AE014297 AY070518 AAF55939.1 AAL47989.1 GGMR01008253 MBY20872.1 CM000160 EDW97930.1 CM000364 EDX13855.1 ABLF02026944 KK855357 PTY22158.1 GL764539 EFZ17500.1 CH954182 EDV54213.1 DQ088697 AAZ06444.1 EEZ99709.1 GGMS01016454 MBY85657.1 CH902617 EDV42569.1 CR381643 KK800603 KFQ28460.1
PCG74375.1 PCG74377.1 PCG74376.1 KQ459605 KPI91907.1 MF319731 ASJ26445.1 JTDY01002142 KOB72015.1 ODYU01005567 SOQ46541.1 AGBW02009564 OWR50415.1 GEZM01061174 JAV70707.1 GEZM01061173 JAV70708.1 APGK01050031 APGK01050032 KB741165 ENN73390.1 GALX01002771 JAB65695.1 KB632352 ERL93296.1 KC282396 AGX25153.1 KQ436033 KOX67532.1 KQ971331 KYB28312.1 KQ978379 KYM94477.1 KQ434978 KZC13020.1 EEZ99710.2 KK107921 EZA47207.1 LBMM01002888 KMQ94231.1 KQ981905 KYN33333.1 GL887695 EGI70400.1 GL438137 EFN69480.1 KQ982576 KYQ54667.1 ADTU01007399 ADTU01007400 ADTU01007401 GL446901 EFN87085.1 KQ976518 KYM82197.1 KQ980903 KYN11595.1 KZ288279 PBC29685.1 KQ414851 KOC60182.1 NEVH01025174 PNF15507.1 PYGN01002482 PSN30415.1 KA645070 AFP59699.1 KK852680 KDR18624.1 CH902619 EDV37879.1 DS235817 EEB17630.1 JRES01001578 KNC21807.1 CM000071 EAL24825.1 CH479181 EDW31549.1 CH940648 EDW59920.1 UFQS01000067 UFQT01000067 SSW98991.1 SSX19373.1 OUUW01000001 SPP73895.1 GFDG01001403 JAV17396.1 GFDG01001404 JAV17395.1 CM000158 EDW90643.1 CP012524 ALC41135.1 CCAG010000868 CH916367 EDW01757.1 CH954179 EDV56189.1 GAKP01017045 JAC41907.1 GDHF01018240 GDHF01012411 GDHF01009786 JAI34074.1 JAI39903.1 JAI42528.1 AE013599 BT004854 AAF58400.1 AAO45210.1 GECZ01021764 JAS48005.1 JXJN01022230 GEBQ01027536 JAT12441.1 CM000362 CM002911 EDX06900.1 KMY93452.1 CH480816 EDW47696.1 GBYB01006313 JAG76080.1 GBXI01002759 JAD11533.1 CH964272 EDW84341.1 GBYB01006311 JAG76078.1 KRG00013.1 KQ971358 KYB26006.1 GAMC01011216 JAB95339.1 GAPW01000673 JAC12925.1 GAPW01000431 JAC13167.1 GAPW01000646 JAC12952.1 JXUM01034905 JXUM01034906 KQ561040 KXJ79927.1 JXUM01051977 JXUM01051978 KQ561713 KXJ77746.1 CH477833 EAT35810.1 AE014297 AY070518 AAF55939.1 AAL47989.1 GGMR01008253 MBY20872.1 CM000160 EDW97930.1 CM000364 EDX13855.1 ABLF02026944 KK855357 PTY22158.1 GL764539 EFZ17500.1 CH954182 EDV54213.1 DQ088697 AAZ06444.1 EEZ99709.1 GGMS01016454 MBY85657.1 CH902617 EDV42569.1 CR381643 KK800603 KFQ28460.1
Proteomes
UP000005204
UP000053240
UP000218220
UP000053268
UP000037510
UP000007151
+ More
UP000192223 UP000019118 UP000030742 UP000053105 UP000007266 UP000078542 UP000076502 UP000053097 UP000036403 UP000078541 UP000007755 UP000000311 UP000075809 UP000002358 UP000005205 UP000008237 UP000078540 UP000078492 UP000005203 UP000242457 UP000053825 UP000235965 UP000245037 UP000095301 UP000027135 UP000007801 UP000009046 UP000037069 UP000078200 UP000001819 UP000008744 UP000008792 UP000268350 UP000002282 UP000092553 UP000092444 UP000095300 UP000001070 UP000008711 UP000092445 UP000091820 UP000000803 UP000092460 UP000092443 UP000000304 UP000001292 UP000192221 UP000007798 UP000069940 UP000249989 UP000008820 UP000007819 UP000079169 UP000000437
UP000192223 UP000019118 UP000030742 UP000053105 UP000007266 UP000078542 UP000076502 UP000053097 UP000036403 UP000078541 UP000007755 UP000000311 UP000075809 UP000002358 UP000005205 UP000008237 UP000078540 UP000078492 UP000005203 UP000242457 UP000053825 UP000235965 UP000245037 UP000095301 UP000027135 UP000007801 UP000009046 UP000037069 UP000078200 UP000001819 UP000008744 UP000008792 UP000268350 UP000002282 UP000092553 UP000092444 UP000095300 UP000001070 UP000008711 UP000092445 UP000091820 UP000000803 UP000092460 UP000092443 UP000000304 UP000001292 UP000192221 UP000007798 UP000069940 UP000249989 UP000008820 UP000007819 UP000079169 UP000000437
Interpro
Gene 3D
ProteinModelPortal
H9ITA1
A0A194QN37
A0A2W1BU08
A0A2A4JQP4
A0A2A4JQR5
A0A2A4JRH9
+ More
A0A194PF51 A0A220K8N7 A0A0L7L9F1 A0A2H1W0E0 A0A212F9J4 A0A1W4XTZ0 A0A1Y1LI55 A0A1Y1LCT5 N6TVR9 V5G6I6 U4USZ4 V9PB47 A0A0N1ISZ6 A0A139WK54 A0A195C0V8 A0A154PPC5 D6WHZ3 A0A026VU03 A0A0J7KVA4 A0A195EYN4 F4W5S4 E2AAN0 A0A151X2K2 K7J8R1 A0A158P2R4 E2BAZ4 A0A151I2S1 A0A195DFB6 A0A088A7I5 A0A2A3ED61 A0A0L7QNG1 A0A2J7PGP5 A0A2P8XEJ0 T1PA99 A0A1I8NB55 A0A067REY5 B3MGW1 E0VW74 A0A0L0BP21 A0A1A9VDD5 Q292N7 B4GD11 B4LM00 A0A336K3J4 A0A3B0IZY0 A0A1L8EFD2 A0A1L8EFK0 B4P4K4 A0A0M4EFT4 A0A1B0F9H4 A0A1I8QCB5 B4J5B5 B3NRN7 A0A1A9ZG99 A0A1A9X3I5 A0A034VIE8 A0A0K8V571 Q7JQR3 A0A1B6FCT0 A0A1B0BXH4 A0A1A9YJI3 A0A1B6KLZ5 B4QE06 B4HQ33 A0A1W4UNU0 A0A0C9PXU2 A0A0A1XLR9 B4NLD8 A0A0C9RHA5 A0A0Q9WVL8 A0A139WDJ7 W8C0Z9 A0A023EU40 A0A023EVQ9 A0A023EVD5 A0A182HDX8 A0A182GB20 A0A1S4FVG8 Q16N96 Q9VD61 A0A2S2NV03 B4PKY1 B4QZB9 J9JRQ3 A0A2R7WRG6 E9IPK0 B3P8P9 Q4FDV9 D6WHZ4 A0A2S2R6H8 A0A3Q0JDK6 B3LVJ9 B0UXN0 A0A091QLP6
A0A194PF51 A0A220K8N7 A0A0L7L9F1 A0A2H1W0E0 A0A212F9J4 A0A1W4XTZ0 A0A1Y1LI55 A0A1Y1LCT5 N6TVR9 V5G6I6 U4USZ4 V9PB47 A0A0N1ISZ6 A0A139WK54 A0A195C0V8 A0A154PPC5 D6WHZ3 A0A026VU03 A0A0J7KVA4 A0A195EYN4 F4W5S4 E2AAN0 A0A151X2K2 K7J8R1 A0A158P2R4 E2BAZ4 A0A151I2S1 A0A195DFB6 A0A088A7I5 A0A2A3ED61 A0A0L7QNG1 A0A2J7PGP5 A0A2P8XEJ0 T1PA99 A0A1I8NB55 A0A067REY5 B3MGW1 E0VW74 A0A0L0BP21 A0A1A9VDD5 Q292N7 B4GD11 B4LM00 A0A336K3J4 A0A3B0IZY0 A0A1L8EFD2 A0A1L8EFK0 B4P4K4 A0A0M4EFT4 A0A1B0F9H4 A0A1I8QCB5 B4J5B5 B3NRN7 A0A1A9ZG99 A0A1A9X3I5 A0A034VIE8 A0A0K8V571 Q7JQR3 A0A1B6FCT0 A0A1B0BXH4 A0A1A9YJI3 A0A1B6KLZ5 B4QE06 B4HQ33 A0A1W4UNU0 A0A0C9PXU2 A0A0A1XLR9 B4NLD8 A0A0C9RHA5 A0A0Q9WVL8 A0A139WDJ7 W8C0Z9 A0A023EU40 A0A023EVQ9 A0A023EVD5 A0A182HDX8 A0A182GB20 A0A1S4FVG8 Q16N96 Q9VD61 A0A2S2NV03 B4PKY1 B4QZB9 J9JRQ3 A0A2R7WRG6 E9IPK0 B3P8P9 Q4FDV9 D6WHZ4 A0A2S2R6H8 A0A3Q0JDK6 B3LVJ9 B0UXN0 A0A091QLP6
PDB
4P2L
E-value=9.05688e-62,
Score=602
Ontologies
GO
PANTHER
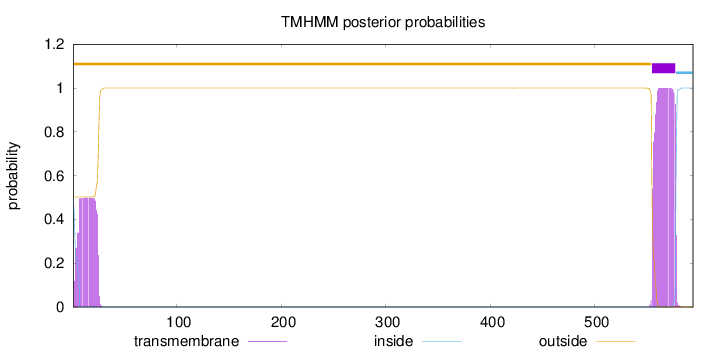
Topology
SignalP
Position: 1 - 19,
Likelihood: 0.952871
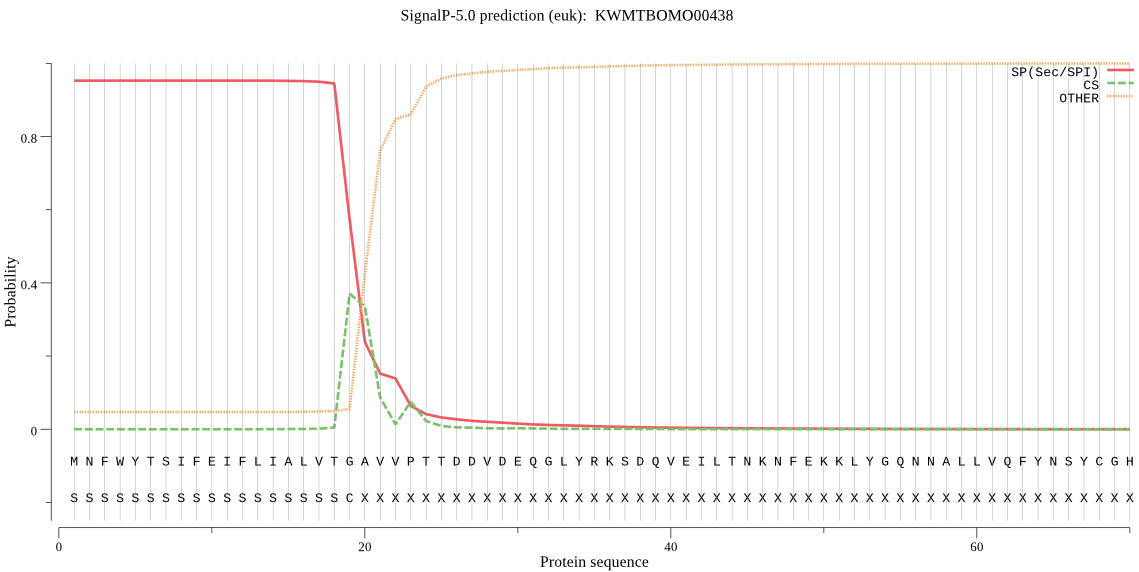
Length:
594
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
32.66204
Exp number, first 60 AAs:
10.45618
Total prob of N-in:
0.49779
POSSIBLE N-term signal
sequence
outside
1 - 554
TMhelix
555 - 577
inside
578 - 594
Population Genetic Test Statistics
Pi
2.325076
Theta
2.36405
Tajima's D
-0.516776
CLR
0.184463
CSRT
0.254937253137343
Interpretation
Uncertain
