Gene
KWMTBOMO00437 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000743
Annotation
PREDICTED:_flotillin-1_isoform_X1_[Bombyx_mori]
Full name
Flotillin-1
Location in the cell
Cytoplasmic Reliability : 2.328
Sequence
CDS
ATGACTTGGGGGTTCGTCACATGTGGCCCTAACGAAGCACTCGTGATTTCAGGATGCTGCTATTCCAAGCCACTCCTGGTCCCTGGTGGTCGAGCCTTTGTATGGCCAGCGATCCAGAGCGTACAACGGATATCTCTAAACACCATGACCCTTCAAGTGGAGTCACCAACTGTCTATACCAGTCAGGGTGTGCCCATTTCGGTTACTGGAATCGCTCAGGTCAAAATTCAAGGTCAGAATTCCGAGATGCTGCTCTCCGCGTGTGAGCAGTTTCTCGGTAAAACGGAGCAAGAGATTCAGCACATAGCCCTGGTGACGCTGGAGGGTCACCAACGTGCTATAATGGGATCTATGACGGTCGAGGAAATATACAAAGACAGAAAAATATTCTCGAAAAAGGTGTTCGAAGTTGCGTCGAGTGATCTAATCAACATGGGAATCACAGTTGTGTCGTATACTTTGAAGGACATCAGAGACGAGGAGGGCTACTTGAAAGCTCTAGGTATGGCTCGTACAGCCGAAGTGAAGCGCGACGCACGTATAGGCGAAGCTGAAGCTCAGGCCGAAGCCAAGATTAAGGAGGCGATGGCTGAGGAGCAACGCATGGCCGCCAGGTTCCTGAACGATACGGAGATCGCTAAGTCTCAGCGGGACTTTGAGCTAAAGAAGGCCGCTTACGACGTAGAAGTTCACACCAAAAAGGCCGAAGCCGAGATGGCCTACGAACTACAAGCTGCCAAGACGAAACAGCGTATCAAAGAGGAACAGATGCAGATAGCGGTCGTGGAGCGAACCCAGGAGATCGCCGTACAGAAGTGGGAAGTACAAAGACGCGAGAAGGAACTGGAAGCCACAATACGCAGACCCGCCGAAGCTGAGAAGTTCAGACTAGAGAAGATAGCGGAAGCGCACAGGCAGAAGACAGTGCTCGAAGCCGAAGCTGAGGCGGAAGCGGTTAAAGTACGAGGAGAGGCGGAAGCATACGCTATAAAAGCAAAGGCCGTTGCTGATGCCGAGCAGATGGCGAAAAAGGCAGAAGCGTGGAAGGAATATGGATCTGCCGCTATGGTGGACATGATGCTGGAAACTCTACCCAAGGTTGCAGCAGAAGTGGCTGCCCCCCTGTCCCAAGCCAGGAAGGTAACTATGGTGTCGTGCGGAGGCGGCGAGGTCGGTGCGGCCAAACTCACCGGTGAAGTATTGAGCATTGTACAGTGCTTACCTGAACTCGTGAAGGGAGTCACCGGAGTCGATATATCCAAGGGTGTTCGCGCACTGTAA
Protein
MTWGFVTCGPNEALVISGCCYSKPLLVPGGRAFVWPAIQSVQRISLNTMTLQVESPTVYTSQGVPISVTGIAQVKIQGQNSEMLLSACEQFLGKTEQEIQHIALVTLEGHQRAIMGSMTVEEIYKDRKIFSKKVFEVASSDLINMGITVVSYTLKDIRDEEGYLKALGMARTAEVKRDARIGEAEAQAEAKIKEAMAEEQRMAARFLNDTEIAKSQRDFELKKAAYDVEVHTKKAEAEMAYELQAAKTKQRIKEEQMQIAVVERTQEIAVQKWEVQRREKELEATIRRPAEAEKFRLEKIAEAHRQKTVLEAEAEAEAVKVRGEAEAYAIKAKAVADAEQMAKKAEAWKEYGSAAMVDMMLETLPKVAAEVAAPLSQARKVTMVSCGGGEVGAAKLTGEVLSIVQCLPELVKGVTGVDISKGVRAL
Summary
Description
May act as a scaffolding protein within caveolar membranes, functionally participating in formation of caveolae or caveolae-like vesicles.
Subunit
Heterooligomeric complex of flotillins 1 and 2 and caveolins 1 and 2.
Similarity
Belongs to the band 7/mec-2 family. Flotillin subfamily.
Belongs to the mitochondrial carrier (TC 2.A.29) family.
Belongs to the mitochondrial carrier (TC 2.A.29) family.
Keywords
Alternative splicing
Cell membrane
Complete proteome
Developmental protein
Membrane
Reference proteome
Feature
chain Flotillin-1
splice variant In isoform Long.
splice variant In isoform Long.
Uniprot
H9IU13
A0A2W1C063
A0A194PFV0
A0A212F9J2
A0A067R5R6
A0A2J7PGP4
+ More
A0A1Q3FTX7 U5EWZ7 A0A182QEA8 A0A2M3Z2T6 A0A2M4A236 A0A182FF53 A0A2M4BNU2 A0A453Z3S4 A0A1Y9IVP9 A0A0K8TSA2 A0A1B6J7Q5 A0A2M3ZGR3 A0A1B6M950 A0A1B0CJH3 W5JNL9 A0A182HDX9 A0A1L8DVS9 A0A1B6G9M7 A0A1I8Q0W3 A0A1B6EG12 W8BIE9 A0A1I8MYU4 A0A1W4UGN5 A0A1Q3FVA1 A0A0P9BR37 A0A3B0JLX0 R4WPP3 Q7QJF7 A0A182RSP4 A0A0R3NMP5 A0A336LHD1 A0A336K0U0 A0A0K8VT48 A0A182K2I2 A0A0K8TY35 N6T011 A0A1I8Q0Y4 A0A1I8MYU0 E0VW73 A0A1W4UGJ6 W8BDL3 D6WVX3 A0A0J9REN1 A0A2R7W6F8 O61491 B3MUP6 A0A3B0J349 A0A0R1DSU8 B4MQW2 A0A0Q5VM84 A0A1Y1MT85 Q290U2 B4GB86 A0A0K8V3B8 A0A0K8W3P4 A0A0A1X4Q5 A0A0Q9XG90 A0A1J1IUH5 A0A0M3QUS5 A0A0P4VXL4 T1I757 A0A0L0BP94 O61491-2 A0A0Q9W2Y1 B4HS60 B4QGW6 B4P7B4 B3NQB1 A0A224XGW3 A0A0T6BAK5 A0A023FAK5 U4UH26 A0A232F1Y4 A0A069DUJ0 A0A0A9W8E3 B4KLK2 B4LM10 A0A2A3EM04 K7J6B6 A0A182KRD4 E2BW52 A0A026WEZ4 B4J5A5 A0A151J6F3 A0A088A9W3 A0A3L8E1A4 A0A151XI90 A0A151IFD8 A0A2M4BPI7 Q16EE7 A0A195FHI3 A0A0M8ZT24 A0A158ND57 A0A195BNR3
A0A1Q3FTX7 U5EWZ7 A0A182QEA8 A0A2M3Z2T6 A0A2M4A236 A0A182FF53 A0A2M4BNU2 A0A453Z3S4 A0A1Y9IVP9 A0A0K8TSA2 A0A1B6J7Q5 A0A2M3ZGR3 A0A1B6M950 A0A1B0CJH3 W5JNL9 A0A182HDX9 A0A1L8DVS9 A0A1B6G9M7 A0A1I8Q0W3 A0A1B6EG12 W8BIE9 A0A1I8MYU4 A0A1W4UGN5 A0A1Q3FVA1 A0A0P9BR37 A0A3B0JLX0 R4WPP3 Q7QJF7 A0A182RSP4 A0A0R3NMP5 A0A336LHD1 A0A336K0U0 A0A0K8VT48 A0A182K2I2 A0A0K8TY35 N6T011 A0A1I8Q0Y4 A0A1I8MYU0 E0VW73 A0A1W4UGJ6 W8BDL3 D6WVX3 A0A0J9REN1 A0A2R7W6F8 O61491 B3MUP6 A0A3B0J349 A0A0R1DSU8 B4MQW2 A0A0Q5VM84 A0A1Y1MT85 Q290U2 B4GB86 A0A0K8V3B8 A0A0K8W3P4 A0A0A1X4Q5 A0A0Q9XG90 A0A1J1IUH5 A0A0M3QUS5 A0A0P4VXL4 T1I757 A0A0L0BP94 O61491-2 A0A0Q9W2Y1 B4HS60 B4QGW6 B4P7B4 B3NQB1 A0A224XGW3 A0A0T6BAK5 A0A023FAK5 U4UH26 A0A232F1Y4 A0A069DUJ0 A0A0A9W8E3 B4KLK2 B4LM10 A0A2A3EM04 K7J6B6 A0A182KRD4 E2BW52 A0A026WEZ4 B4J5A5 A0A151J6F3 A0A088A9W3 A0A3L8E1A4 A0A151XI90 A0A151IFD8 A0A2M4BPI7 Q16EE7 A0A195FHI3 A0A0M8ZT24 A0A158ND57 A0A195BNR3
Pubmed
19121390
28756777
26354079
22118469
24845553
26369729
+ More
20920257 23761445 26483478 24495485 25315136 17994087 23691247 12364791 14747013 17210077 15632085 23537049 20566863 18362917 19820115 22936249 9573373 10731132 12537572 12537569 17550304 28004739 25830018 27129103 26108605 25474469 28648823 26334808 25401762 26823975 20075255 20966253 20798317 24508170 30249741 17510324 21347285
20920257 23761445 26483478 24495485 25315136 17994087 23691247 12364791 14747013 17210077 15632085 23537049 20566863 18362917 19820115 22936249 9573373 10731132 12537572 12537569 17550304 28004739 25830018 27129103 26108605 25474469 28648823 26334808 25401762 26823975 20075255 20966253 20798317 24508170 30249741 17510324 21347285
EMBL
BABH01007335
KZ149901
PZC78500.1
KQ459605
KPI91908.1
AGBW02009564
+ More
OWR50416.1 KK852680 KDR18625.1 NEVH01025174 PNF15505.1 GFDL01004030 JAV31015.1 GANO01001196 JAB58675.1 AXCN02000377 GGFM01002080 MBW22831.1 GGFK01001491 MBW34812.1 GGFJ01005609 MBW54750.1 GDAI01000369 JAI17234.1 GECU01037022 GECU01030300 GECU01022134 GECU01012499 JAS70684.1 JAS77406.1 JAS85572.1 JAS95207.1 GGFM01006932 MBW27683.1 GEBQ01007522 JAT32455.1 AJWK01014645 AJWK01014646 ADMH02000549 ETN65967.1 JXUM01034908 JXUM01034909 JXUM01034910 KQ561040 KXJ79928.1 GFDF01003540 JAV10544.1 GECZ01010658 JAS59111.1 GEDC01000443 JAS36855.1 GAMC01009772 JAB96783.1 GFDL01003521 JAV31524.1 CH902624 KPU74210.1 OUUW01000001 SPP73641.1 AK417636 BAN20851.1 AAAB01008807 EAA04642.4 CM000071 KRT02028.1 UFQS01000047 UFQS01003939 UFQT01000047 UFQT01003939 SSW98527.1 SSX16045.1 SSX18913.1 SSW98526.1 SSX18912.1 GDHF01010286 JAI42028.1 GDHF01033319 JAI18995.1 APGK01050028 APGK01050029 APGK01050030 KB741165 KB632352 ENN73389.1 ERL93295.1 DS235817 EEB17629.1 GAMC01009773 JAB96782.1 KQ971358 EFA08627.2 CM002911 KMY94029.1 KK854254 PTY13795.1 AF044734 AE013599 AY058794 EDV32961.1 SPP73642.1 CM000158 KRK00294.1 CH963849 EDW74501.1 CH954179 KQS62535.1 GEZM01021631 JAV88893.1 EAL25270.2 CH479181 EDW32188.1 GDHF01018897 JAI33417.1 GDHF01006610 JAI45704.1 GBXI01008471 JAD05821.1 CH933808 KRG04212.1 CVRI01000059 CRL03354.1 CP012524 ALC41143.1 GDKW01001193 JAI55402.1 ACPB03000761 JRES01001578 KNC21841.1 CH940648 KRF79158.1 CH480816 EDW48007.1 CM000362 EDX07229.1 KMY94028.1 EDW92059.1 EDV55887.1 GFTR01006162 JAW10264.1 LJIG01002564 KRT84376.1 GBBI01000181 JAC18531.1 ERL93294.1 NNAY01001263 OXU24572.1 GBGD01001542 JAC87347.1 GBHO01039540 GBHO01039535 GDHC01021038 JAG04064.1 JAG04069.1 JAP97590.1 EDW08638.1 EDW59930.1 KZ288212 PBC32833.1 GL451091 EFN80084.1 KK107260 EZA54251.1 CH916367 EDW01747.1 KQ979878 KYN18692.1 QOIP01000001 RLU26243.1 KQ982116 KYQ59930.1 KQ977796 KYM99616.1 GGFJ01005855 MBW54996.1 CH478807 EAT32605.1 KQ981560 KYN39848.1 KQ435891 KOX69406.1 ADTU01012403 KQ976438 KYM86873.1
OWR50416.1 KK852680 KDR18625.1 NEVH01025174 PNF15505.1 GFDL01004030 JAV31015.1 GANO01001196 JAB58675.1 AXCN02000377 GGFM01002080 MBW22831.1 GGFK01001491 MBW34812.1 GGFJ01005609 MBW54750.1 GDAI01000369 JAI17234.1 GECU01037022 GECU01030300 GECU01022134 GECU01012499 JAS70684.1 JAS77406.1 JAS85572.1 JAS95207.1 GGFM01006932 MBW27683.1 GEBQ01007522 JAT32455.1 AJWK01014645 AJWK01014646 ADMH02000549 ETN65967.1 JXUM01034908 JXUM01034909 JXUM01034910 KQ561040 KXJ79928.1 GFDF01003540 JAV10544.1 GECZ01010658 JAS59111.1 GEDC01000443 JAS36855.1 GAMC01009772 JAB96783.1 GFDL01003521 JAV31524.1 CH902624 KPU74210.1 OUUW01000001 SPP73641.1 AK417636 BAN20851.1 AAAB01008807 EAA04642.4 CM000071 KRT02028.1 UFQS01000047 UFQS01003939 UFQT01000047 UFQT01003939 SSW98527.1 SSX16045.1 SSX18913.1 SSW98526.1 SSX18912.1 GDHF01010286 JAI42028.1 GDHF01033319 JAI18995.1 APGK01050028 APGK01050029 APGK01050030 KB741165 KB632352 ENN73389.1 ERL93295.1 DS235817 EEB17629.1 GAMC01009773 JAB96782.1 KQ971358 EFA08627.2 CM002911 KMY94029.1 KK854254 PTY13795.1 AF044734 AE013599 AY058794 EDV32961.1 SPP73642.1 CM000158 KRK00294.1 CH963849 EDW74501.1 CH954179 KQS62535.1 GEZM01021631 JAV88893.1 EAL25270.2 CH479181 EDW32188.1 GDHF01018897 JAI33417.1 GDHF01006610 JAI45704.1 GBXI01008471 JAD05821.1 CH933808 KRG04212.1 CVRI01000059 CRL03354.1 CP012524 ALC41143.1 GDKW01001193 JAI55402.1 ACPB03000761 JRES01001578 KNC21841.1 CH940648 KRF79158.1 CH480816 EDW48007.1 CM000362 EDX07229.1 KMY94028.1 EDW92059.1 EDV55887.1 GFTR01006162 JAW10264.1 LJIG01002564 KRT84376.1 GBBI01000181 JAC18531.1 ERL93294.1 NNAY01001263 OXU24572.1 GBGD01001542 JAC87347.1 GBHO01039540 GBHO01039535 GDHC01021038 JAG04064.1 JAG04069.1 JAP97590.1 EDW08638.1 EDW59930.1 KZ288212 PBC32833.1 GL451091 EFN80084.1 KK107260 EZA54251.1 CH916367 EDW01747.1 KQ979878 KYN18692.1 QOIP01000001 RLU26243.1 KQ982116 KYQ59930.1 KQ977796 KYM99616.1 GGFJ01005855 MBW54996.1 CH478807 EAT32605.1 KQ981560 KYN39848.1 KQ435891 KOX69406.1 ADTU01012403 KQ976438 KYM86873.1
Proteomes
UP000005204
UP000053268
UP000007151
UP000027135
UP000235965
UP000075886
+ More
UP000069272 UP000076407 UP000075920 UP000092461 UP000000673 UP000069940 UP000249989 UP000095300 UP000095301 UP000192221 UP000007801 UP000268350 UP000007062 UP000075900 UP000001819 UP000075881 UP000019118 UP000030742 UP000009046 UP000007266 UP000000803 UP000002282 UP000007798 UP000008711 UP000008744 UP000009192 UP000183832 UP000092553 UP000015103 UP000037069 UP000008792 UP000001292 UP000000304 UP000215335 UP000242457 UP000002358 UP000075882 UP000008237 UP000053097 UP000001070 UP000078492 UP000005203 UP000279307 UP000075809 UP000078542 UP000008820 UP000078541 UP000053105 UP000005205 UP000078540
UP000069272 UP000076407 UP000075920 UP000092461 UP000000673 UP000069940 UP000249989 UP000095300 UP000095301 UP000192221 UP000007801 UP000268350 UP000007062 UP000075900 UP000001819 UP000075881 UP000019118 UP000030742 UP000009046 UP000007266 UP000000803 UP000002282 UP000007798 UP000008711 UP000008744 UP000009192 UP000183832 UP000092553 UP000015103 UP000037069 UP000008792 UP000001292 UP000000304 UP000215335 UP000242457 UP000002358 UP000075882 UP000008237 UP000053097 UP000001070 UP000078492 UP000005203 UP000279307 UP000075809 UP000078542 UP000008820 UP000078541 UP000053105 UP000005205 UP000078540
Pfam
Interpro
IPR001107
Band_7
+ More
IPR031905 Flotillin_C
IPR027705 Flotillin_fam
IPR036013 Band_7/SPFH_dom_sf
IPR010756 Tls1
IPR010675 Bin3_C
IPR029063 SAM-dependent_MTases
IPR024160 BIN3_SAM-bd_dom
IPR041698 Methyltransf_25
IPR002113 Aden_trnslctor
IPR002067 Mit_carrier
IPR023395 Mt_carrier_dom_sf
IPR018108 Mitochondrial_sb/sol_carrier
IPR031905 Flotillin_C
IPR027705 Flotillin_fam
IPR036013 Band_7/SPFH_dom_sf
IPR010756 Tls1
IPR010675 Bin3_C
IPR029063 SAM-dependent_MTases
IPR024160 BIN3_SAM-bd_dom
IPR041698 Methyltransf_25
IPR002113 Aden_trnslctor
IPR002067 Mit_carrier
IPR023395 Mt_carrier_dom_sf
IPR018108 Mitochondrial_sb/sol_carrier
Gene 3D
ProteinModelPortal
H9IU13
A0A2W1C063
A0A194PFV0
A0A212F9J2
A0A067R5R6
A0A2J7PGP4
+ More
A0A1Q3FTX7 U5EWZ7 A0A182QEA8 A0A2M3Z2T6 A0A2M4A236 A0A182FF53 A0A2M4BNU2 A0A453Z3S4 A0A1Y9IVP9 A0A0K8TSA2 A0A1B6J7Q5 A0A2M3ZGR3 A0A1B6M950 A0A1B0CJH3 W5JNL9 A0A182HDX9 A0A1L8DVS9 A0A1B6G9M7 A0A1I8Q0W3 A0A1B6EG12 W8BIE9 A0A1I8MYU4 A0A1W4UGN5 A0A1Q3FVA1 A0A0P9BR37 A0A3B0JLX0 R4WPP3 Q7QJF7 A0A182RSP4 A0A0R3NMP5 A0A336LHD1 A0A336K0U0 A0A0K8VT48 A0A182K2I2 A0A0K8TY35 N6T011 A0A1I8Q0Y4 A0A1I8MYU0 E0VW73 A0A1W4UGJ6 W8BDL3 D6WVX3 A0A0J9REN1 A0A2R7W6F8 O61491 B3MUP6 A0A3B0J349 A0A0R1DSU8 B4MQW2 A0A0Q5VM84 A0A1Y1MT85 Q290U2 B4GB86 A0A0K8V3B8 A0A0K8W3P4 A0A0A1X4Q5 A0A0Q9XG90 A0A1J1IUH5 A0A0M3QUS5 A0A0P4VXL4 T1I757 A0A0L0BP94 O61491-2 A0A0Q9W2Y1 B4HS60 B4QGW6 B4P7B4 B3NQB1 A0A224XGW3 A0A0T6BAK5 A0A023FAK5 U4UH26 A0A232F1Y4 A0A069DUJ0 A0A0A9W8E3 B4KLK2 B4LM10 A0A2A3EM04 K7J6B6 A0A182KRD4 E2BW52 A0A026WEZ4 B4J5A5 A0A151J6F3 A0A088A9W3 A0A3L8E1A4 A0A151XI90 A0A151IFD8 A0A2M4BPI7 Q16EE7 A0A195FHI3 A0A0M8ZT24 A0A158ND57 A0A195BNR3
A0A1Q3FTX7 U5EWZ7 A0A182QEA8 A0A2M3Z2T6 A0A2M4A236 A0A182FF53 A0A2M4BNU2 A0A453Z3S4 A0A1Y9IVP9 A0A0K8TSA2 A0A1B6J7Q5 A0A2M3ZGR3 A0A1B6M950 A0A1B0CJH3 W5JNL9 A0A182HDX9 A0A1L8DVS9 A0A1B6G9M7 A0A1I8Q0W3 A0A1B6EG12 W8BIE9 A0A1I8MYU4 A0A1W4UGN5 A0A1Q3FVA1 A0A0P9BR37 A0A3B0JLX0 R4WPP3 Q7QJF7 A0A182RSP4 A0A0R3NMP5 A0A336LHD1 A0A336K0U0 A0A0K8VT48 A0A182K2I2 A0A0K8TY35 N6T011 A0A1I8Q0Y4 A0A1I8MYU0 E0VW73 A0A1W4UGJ6 W8BDL3 D6WVX3 A0A0J9REN1 A0A2R7W6F8 O61491 B3MUP6 A0A3B0J349 A0A0R1DSU8 B4MQW2 A0A0Q5VM84 A0A1Y1MT85 Q290U2 B4GB86 A0A0K8V3B8 A0A0K8W3P4 A0A0A1X4Q5 A0A0Q9XG90 A0A1J1IUH5 A0A0M3QUS5 A0A0P4VXL4 T1I757 A0A0L0BP94 O61491-2 A0A0Q9W2Y1 B4HS60 B4QGW6 B4P7B4 B3NQB1 A0A224XGW3 A0A0T6BAK5 A0A023FAK5 U4UH26 A0A232F1Y4 A0A069DUJ0 A0A0A9W8E3 B4KLK2 B4LM10 A0A2A3EM04 K7J6B6 A0A182KRD4 E2BW52 A0A026WEZ4 B4J5A5 A0A151J6F3 A0A088A9W3 A0A3L8E1A4 A0A151XI90 A0A151IFD8 A0A2M4BPI7 Q16EE7 A0A195FHI3 A0A0M8ZT24 A0A158ND57 A0A195BNR3
PDB
1WIN
E-value=3.36274e-29,
Score=320
Ontologies
PANTHER
Topology
Subcellular location
Cell membrane
Membrane
Caveola
Membrane
Caveola
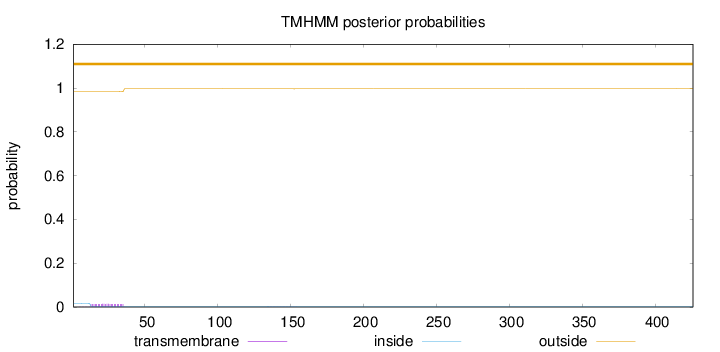
Length:
426
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.331340000000001
Exp number, first 60 AAs:
0.31649
Total prob of N-in:
0.01672
outside
1 - 426
Population Genetic Test Statistics
Pi
0.920425
Theta
2.297353
Tajima's D
-2.224076
CLR
0.053056
CSRT
0.00129993500324984
Interpretation
Uncertain
