Gene
KWMTBOMO00435
Pre Gene Modal
BGIBMGA000742
Annotation
PREDICTED:_RING_finger_and_SPRY_domain-containing_protein_1-like_isoform_X1_[Bombyx_mori]
Full name
RING finger and SPRY domain-containing protein 1
Location in the cell
Extracellular Reliability : 1.346 PlasmaMembrane Reliability : 1.59
Sequence
CDS
ATGATAGCGATGCTGGGCGATGAAGAGTCACCGGTTCTGCTCGTCAAACTGCACGTGATCGCTGACAAAGAAGAGGGATGGCTGCAAGTCGTCTCGTCAATGGTCAACGTGATACCACTCGACAATCCGCTGGGACCGTCGGCGATCACCATAGTCTTTGATGATTGCCCGCTACCCTCCAGAGACTCTGTCATAAAGCTAACGAAACTGTTCCGGCTGTCGAGGCAACGTGCACTCTCGAACGCCCTCGACGTGAAGATGAAACGCAATATGTGCGTTGTCCTCGGGTGTATTGCCGAGAAACTGGCGGGCCCGAACAGCGTCGCTGTACTCACCGAAGAGACGCTCGACTTTCTCCTTGCATTTCTGGACACCAACTTCGAGCCGTGCATAGTGCTCTTCGCACTAATCGCCCTTGAGAAGTTCGCTCACACCACCGAAAACAAGCTGGTGATAAGAAGCCGCTTCGACAAAGAAGACGTGAATCCACTTCTGAAACTCGAGAAACTTGCCAATAATGAAGATTATGTCTGGAGGCAGGTCGGCTTTTGTGCTCAGTGGGCCCTCGACAACTTGTTTGTAATCGAGAACAGGCCTCTGTCGTACGAGACCGTCGACATGACTCCTATAAACGTGATCCTGAATTCGCATGACGTCAGCGAATACCTGAAGATATCTTGCAACGGCCTAGAGGCGCGCTGCGATTCGTACTCGTTTGAAAGCGTACGCTGCACTTTCCAGGTGGACAATGGCTGTTGGTACTACGAGGTGACGATCGTCACTCCTGGCGTCATGCAGATCGGGTGGGCCACCAAGAACAGCCATTTTCTGAACCATGAGGGCTACGGTATCGGCGACGATTTGTATTCGCTCTCGTACGACGGGTGCAGGAAGTTGATTTGGTACAAGGCCAAGCCCGCTTCGGTCCAAGATCTACCGGAGTGGCAGCCCGGAGACATACTCGGCTGTCTCATCGATCTGAACACGCGCGAGGCAATATTCTCCCTCAACGGGCGCTGTCTTACACCGTGCACCGAAATCTTCGAGACAACCAGGTGCTGCAAACAAGGTTTCTTCGCTGCGGCTAGCTTCATGGCGTTCCAGCAGTGCCGATTCAATTTCGGTCATTTGCCGTTTTCGTTTCCTCCAACTGACAGGCCCTTCATGGCCTTCAACGATTACGCGCGTCTCACCGACGAAGAGAAAAAGGTGCTACCTCGCCGCTACTACCTGGAGAAGGTGAGGAATGCTAGCATCCCCGAAAACGCCTGCACTCTTTGCTACGATGCCGTCGCGGACTGCACCTTGGAGCCATGCTTCCACAAGGGATTCTGCTCGAAATGCGCTGCTCAGCTCCGCGAGTGCCCGTTATGCCGCGCTCCGATACTGAACGTGAAGCGCGACATATGA
Protein
MIAMLGDEESPVLLVKLHVIADKEEGWLQVVSSMVNVIPLDNPLGPSAITIVFDDCPLPSRDSVIKLTKLFRLSRQRALSNALDVKMKRNMCVVLGCIAEKLAGPNSVAVLTEETLDFLLAFLDTNFEPCIVLFALIALEKFAHTTENKLVIRSRFDKEDVNPLLKLEKLANNEDYVWRQVGFCAQWALDNLFVIENRPLSYETVDMTPINVILNSHDVSEYLKISCNGLEARCDSYSFESVRCTFQVDNGCWYYEVTIVTPGVMQIGWATKNSHFLNHEGYGIGDDLYSLSYDGCRKLIWYKAKPASVQDLPEWQPGDILGCLIDLNTREAIFSLNGRCLTPCTEIFETTRCCKQGFFAAASFMAFQQCRFNFGHLPFSFPPTDRPFMAFNDYARLTDEEKKVLPRRYYLEKVRNASIPENACTLCYDAVADCTLEPCFHKGFCSKCAAQLRECPLCRAPILNVKRDI
Summary
Keywords
Alternative splicing
Complete proteome
Disease mutation
Dwarfism
Glycoprotein
Mental retardation
Metal-binding
Phosphoprotein
Reference proteome
Secreted
Signal
Zinc
Zinc-finger
Feature
chain RING finger and SPRY domain-containing protein 1
splice variant In isoform 2.
sequence variant In SEMDFA; dbSNP:rs864309652.
splice variant In isoform 2.
sequence variant In SEMDFA; dbSNP:rs864309652.
Uniprot
H9IU12
A0A212F9K9
A0A2A4JLR4
A0A2H1V7T4
A0A195D8I4
A0A087ZTN2
+ More
E2A5D9 F4WU09 A0A151WWB8 A0A158P114 E2BTH6 A0A195DKG7 A0A026VU71 E9IE86 A0A2J7QJA0 A0A151JUR1 A0A0C9PSP3 A0A0L7QXQ5 A0A310S5F5 A0A232F4I1 A0A0M9A0T5 A0A154P927 A0A2W1BXX5 A0A1Y1M940 A0A2A3EHF8 K7IW25 A0A224Z6A8 A0A131YHF9 A0A131X8M0 L7M7R1 A0A1W4WNC8 E0W1N9 T1IHI8 A0A0P4W5Y0 A0A139WM94 A0A084W854 V5HK78 A0A1B6C6Q6 A0A2R5LBY4 A0A0P4XG14 A0A0P5BBS5 A0A0P5KAI0 A0A1Q3F9D4 A0A293LCM5 A0A182TSS6 A0A1Q3F995 A0A182W705 A0A182JGC5 E9HG27 A0A182R105 A0A1Q3F9D7 A0A182NUG3 A0A1S4GXE5 A0A182HUL7 A0A182L1D3 A0A182WU01 A0A182VHE8 Q17FP0 A0A182JPP5 B0WCT8 A0A182PPL0 W5J8V1 A0A182RCS6 A0A069DVB3 A0A182YQX9 A0A023F3S7 A0A0V0G6F2 A0A1B6I9K8 A0A1B6LFK8 A0A226DPH5 A0A182MGX3 A0A336K6M9 A0A182SCG8 A0A146LYJ7 N6U7R3 U4U011 A0A0P5S2K0 H3B0T0 A0A1D2MGZ7 A0A091R5P2 A0A2Y9STG5 A0A383YXL4 A0A2U3V077 A0A340Y961 A0A0F7YZC5 V8NUH3 A0A2Y9LPD4 A0A094L069 A0A2K5MCV3 A0A2R8ZFC6 H2QB64 A0A024R6U0 Q96DX4 A0A2J8VZ28 Q5R881 Q95LP3 G7Q173 G7NPT9 H0VUL8 Q28E18
E2A5D9 F4WU09 A0A151WWB8 A0A158P114 E2BTH6 A0A195DKG7 A0A026VU71 E9IE86 A0A2J7QJA0 A0A151JUR1 A0A0C9PSP3 A0A0L7QXQ5 A0A310S5F5 A0A232F4I1 A0A0M9A0T5 A0A154P927 A0A2W1BXX5 A0A1Y1M940 A0A2A3EHF8 K7IW25 A0A224Z6A8 A0A131YHF9 A0A131X8M0 L7M7R1 A0A1W4WNC8 E0W1N9 T1IHI8 A0A0P4W5Y0 A0A139WM94 A0A084W854 V5HK78 A0A1B6C6Q6 A0A2R5LBY4 A0A0P4XG14 A0A0P5BBS5 A0A0P5KAI0 A0A1Q3F9D4 A0A293LCM5 A0A182TSS6 A0A1Q3F995 A0A182W705 A0A182JGC5 E9HG27 A0A182R105 A0A1Q3F9D7 A0A182NUG3 A0A1S4GXE5 A0A182HUL7 A0A182L1D3 A0A182WU01 A0A182VHE8 Q17FP0 A0A182JPP5 B0WCT8 A0A182PPL0 W5J8V1 A0A182RCS6 A0A069DVB3 A0A182YQX9 A0A023F3S7 A0A0V0G6F2 A0A1B6I9K8 A0A1B6LFK8 A0A226DPH5 A0A182MGX3 A0A336K6M9 A0A182SCG8 A0A146LYJ7 N6U7R3 U4U011 A0A0P5S2K0 H3B0T0 A0A1D2MGZ7 A0A091R5P2 A0A2Y9STG5 A0A383YXL4 A0A2U3V077 A0A340Y961 A0A0F7YZC5 V8NUH3 A0A2Y9LPD4 A0A094L069 A0A2K5MCV3 A0A2R8ZFC6 H2QB64 A0A024R6U0 Q96DX4 A0A2J8VZ28 Q5R881 Q95LP3 G7Q173 G7NPT9 H0VUL8 Q28E18
Pubmed
19121390
22118469
20798317
21719571
21347285
24508170
+ More
30249741 21282665 28648823 28756777 28004739 20075255 28797301 26830274 28049606 25576852 20566863 18362917 19820115 24438588 25765539 21292972 12364791 20966253 17510324 20920257 23761445 26334808 25244985 25474469 26823975 23537049 9215903 27289101 24297900 22722832 16136131 11181995 11853319 12975309 14702039 15489334 17974005 24275569 26365341 12498619 22002653 25319552 21993624
30249741 21282665 28648823 28756777 28004739 20075255 28797301 26830274 28049606 25576852 20566863 18362917 19820115 24438588 25765539 21292972 12364791 20966253 17510324 20920257 23761445 26334808 25244985 25474469 26823975 23537049 9215903 27289101 24297900 22722832 16136131 11181995 11853319 12975309 14702039 15489334 17974005 24275569 26365341 12498619 22002653 25319552 21993624
EMBL
BABH01007321
AGBW02009564
OWR50417.1
NWSH01001050
PCG72921.1
ODYU01000932
+ More
SOQ36442.1 KQ976749 KYN08744.1 GL436921 EFN71361.1 GL888345 EGI62366.1 KQ982691 KYQ52183.1 ADTU01005953 GL450410 EFN81004.1 KQ980765 KYN13332.1 KK107921 QOIP01000001 EZA47190.1 RLU27106.1 GL762567 EFZ21114.1 NEVH01013559 PNF28642.1 KQ981732 KYN36594.1 GBYB01004358 JAG74125.1 KQ414699 KOC63393.1 KQ770168 OAD52697.1 NNAY01001039 OXU25388.1 KQ435797 KOX73499.1 KQ434846 KZC08409.1 KZ149901 PZC78505.1 GEZM01037071 JAV82214.1 KZ288254 PBC30904.1 GFPF01011595 MAA22741.1 GEDV01010190 JAP78367.1 GEFH01004727 JAP63854.1 GACK01004974 JAA60060.1 DS235873 EEB19621.1 JH429910 GDRN01080938 JAI62108.1 KQ971318 KYB28945.1 ATLV01021363 ATLV01021364 KE525317 KFB46398.1 GANP01010570 JAB73898.1 GEDC01028214 JAS09084.1 GGLE01002839 MBY06965.1 GDIP01243206 JAI80195.1 GDIP01187430 JAJ35972.1 GDIQ01188106 GDIP01061198 LRGB01003375 JAK63619.1 JAM42517.1 KZS03011.1 GFDL01010916 JAV24129.1 GFWV01000851 MAA25581.1 GFDL01010900 JAV24145.1 GL732638 EFX69331.1 AXCN02000761 GFDL01010908 JAV24137.1 AAAB01008964 APCN01002465 CH477269 EAT45384.2 DS231890 EDS43814.1 ADMH02001944 ETN60386.1 GBGD01001247 JAC87642.1 GBBI01003148 JAC15564.1 GECL01002504 JAP03620.1 GECU01024108 JAS83598.1 GEBQ01017504 GEBQ01007933 JAT22473.1 JAT32044.1 LNIX01000014 OXA46928.1 AXCM01010210 UFQS01000152 UFQT01000152 SSX00554.1 SSX20934.1 GDHC01005951 JAQ12678.1 APGK01039256 KB740969 ENN76686.1 KB631730 ERL85638.1 GDIQ01093194 JAL58532.1 AFYH01081251 AFYH01081252 AFYH01081253 AFYH01081254 AFYH01081255 AFYH01081256 LJIJ01001280 ODM92278.1 KK809558 KFQ34557.1 GBEW01000820 JAI09545.1 AZIM01001801 ETE65720.1 KL348830 KFZ57554.1 AJFE02083854 AJFE02083855 AJFE02083856 AJFE02083857 AJFE02083858 AJFE02083859 AJFE02083860 AACZ04057487 GABC01010674 GABF01009998 GABD01010852 GABE01011029 GABE01011028 NBAG03000242 JAA00664.1 JAA12147.1 JAA22248.1 JAA33710.1 PNI63784.1 PNI63785.1 CH471092 EAW82911.1 AB075852 AY358548 AK172845 BC013173 AL834402 NDHI03003406 PNJ62756.1 PNJ62758.1 CR859873 AB072745 AQIA01043010 AQIA01043011 AQIA01043012 AQIA01043013 AQIA01043014 AQIA01043015 CM001295 EHH60411.1 JU321980 JU473337 JV044553 CM001272 AFE65736.1 AFH30141.1 AFI34624.1 EHH31691.1 AAKN02026944 BC123024 CR848493 AAI23025.1 CAJ81703.1
SOQ36442.1 KQ976749 KYN08744.1 GL436921 EFN71361.1 GL888345 EGI62366.1 KQ982691 KYQ52183.1 ADTU01005953 GL450410 EFN81004.1 KQ980765 KYN13332.1 KK107921 QOIP01000001 EZA47190.1 RLU27106.1 GL762567 EFZ21114.1 NEVH01013559 PNF28642.1 KQ981732 KYN36594.1 GBYB01004358 JAG74125.1 KQ414699 KOC63393.1 KQ770168 OAD52697.1 NNAY01001039 OXU25388.1 KQ435797 KOX73499.1 KQ434846 KZC08409.1 KZ149901 PZC78505.1 GEZM01037071 JAV82214.1 KZ288254 PBC30904.1 GFPF01011595 MAA22741.1 GEDV01010190 JAP78367.1 GEFH01004727 JAP63854.1 GACK01004974 JAA60060.1 DS235873 EEB19621.1 JH429910 GDRN01080938 JAI62108.1 KQ971318 KYB28945.1 ATLV01021363 ATLV01021364 KE525317 KFB46398.1 GANP01010570 JAB73898.1 GEDC01028214 JAS09084.1 GGLE01002839 MBY06965.1 GDIP01243206 JAI80195.1 GDIP01187430 JAJ35972.1 GDIQ01188106 GDIP01061198 LRGB01003375 JAK63619.1 JAM42517.1 KZS03011.1 GFDL01010916 JAV24129.1 GFWV01000851 MAA25581.1 GFDL01010900 JAV24145.1 GL732638 EFX69331.1 AXCN02000761 GFDL01010908 JAV24137.1 AAAB01008964 APCN01002465 CH477269 EAT45384.2 DS231890 EDS43814.1 ADMH02001944 ETN60386.1 GBGD01001247 JAC87642.1 GBBI01003148 JAC15564.1 GECL01002504 JAP03620.1 GECU01024108 JAS83598.1 GEBQ01017504 GEBQ01007933 JAT22473.1 JAT32044.1 LNIX01000014 OXA46928.1 AXCM01010210 UFQS01000152 UFQT01000152 SSX00554.1 SSX20934.1 GDHC01005951 JAQ12678.1 APGK01039256 KB740969 ENN76686.1 KB631730 ERL85638.1 GDIQ01093194 JAL58532.1 AFYH01081251 AFYH01081252 AFYH01081253 AFYH01081254 AFYH01081255 AFYH01081256 LJIJ01001280 ODM92278.1 KK809558 KFQ34557.1 GBEW01000820 JAI09545.1 AZIM01001801 ETE65720.1 KL348830 KFZ57554.1 AJFE02083854 AJFE02083855 AJFE02083856 AJFE02083857 AJFE02083858 AJFE02083859 AJFE02083860 AACZ04057487 GABC01010674 GABF01009998 GABD01010852 GABE01011029 GABE01011028 NBAG03000242 JAA00664.1 JAA12147.1 JAA22248.1 JAA33710.1 PNI63784.1 PNI63785.1 CH471092 EAW82911.1 AB075852 AY358548 AK172845 BC013173 AL834402 NDHI03003406 PNJ62756.1 PNJ62758.1 CR859873 AB072745 AQIA01043010 AQIA01043011 AQIA01043012 AQIA01043013 AQIA01043014 AQIA01043015 CM001295 EHH60411.1 JU321980 JU473337 JV044553 CM001272 AFE65736.1 AFH30141.1 AFI34624.1 EHH31691.1 AAKN02026944 BC123024 CR848493 AAI23025.1 CAJ81703.1
Proteomes
UP000005204
UP000007151
UP000218220
UP000078542
UP000005203
UP000000311
+ More
UP000007755 UP000075809 UP000005205 UP000008237 UP000078492 UP000053097 UP000279307 UP000235965 UP000078541 UP000053825 UP000215335 UP000053105 UP000076502 UP000242457 UP000002358 UP000192223 UP000009046 UP000007266 UP000030765 UP000076858 UP000075902 UP000075920 UP000075880 UP000000305 UP000075886 UP000075884 UP000075840 UP000075882 UP000076407 UP000075903 UP000008820 UP000075881 UP000002320 UP000075885 UP000000673 UP000075900 UP000076408 UP000198287 UP000075883 UP000075901 UP000019118 UP000030742 UP000008672 UP000094527 UP000248484 UP000261681 UP000245320 UP000265300 UP000248483 UP000233060 UP000240080 UP000002277 UP000005640 UP000001595 UP000233100 UP000009130 UP000005447
UP000007755 UP000075809 UP000005205 UP000008237 UP000078492 UP000053097 UP000279307 UP000235965 UP000078541 UP000053825 UP000215335 UP000053105 UP000076502 UP000242457 UP000002358 UP000192223 UP000009046 UP000007266 UP000030765 UP000076858 UP000075902 UP000075920 UP000075880 UP000000305 UP000075886 UP000075884 UP000075840 UP000075882 UP000076407 UP000075903 UP000008820 UP000075881 UP000002320 UP000075885 UP000000673 UP000075900 UP000076408 UP000198287 UP000075883 UP000075901 UP000019118 UP000030742 UP000008672 UP000094527 UP000248484 UP000261681 UP000245320 UP000265300 UP000248483 UP000233060 UP000240080 UP000002277 UP000005640 UP000001595 UP000233100 UP000009130 UP000005447
Pfam
PF00622 SPRY
Interpro
Gene 3D
ProteinModelPortal
H9IU12
A0A212F9K9
A0A2A4JLR4
A0A2H1V7T4
A0A195D8I4
A0A087ZTN2
+ More
E2A5D9 F4WU09 A0A151WWB8 A0A158P114 E2BTH6 A0A195DKG7 A0A026VU71 E9IE86 A0A2J7QJA0 A0A151JUR1 A0A0C9PSP3 A0A0L7QXQ5 A0A310S5F5 A0A232F4I1 A0A0M9A0T5 A0A154P927 A0A2W1BXX5 A0A1Y1M940 A0A2A3EHF8 K7IW25 A0A224Z6A8 A0A131YHF9 A0A131X8M0 L7M7R1 A0A1W4WNC8 E0W1N9 T1IHI8 A0A0P4W5Y0 A0A139WM94 A0A084W854 V5HK78 A0A1B6C6Q6 A0A2R5LBY4 A0A0P4XG14 A0A0P5BBS5 A0A0P5KAI0 A0A1Q3F9D4 A0A293LCM5 A0A182TSS6 A0A1Q3F995 A0A182W705 A0A182JGC5 E9HG27 A0A182R105 A0A1Q3F9D7 A0A182NUG3 A0A1S4GXE5 A0A182HUL7 A0A182L1D3 A0A182WU01 A0A182VHE8 Q17FP0 A0A182JPP5 B0WCT8 A0A182PPL0 W5J8V1 A0A182RCS6 A0A069DVB3 A0A182YQX9 A0A023F3S7 A0A0V0G6F2 A0A1B6I9K8 A0A1B6LFK8 A0A226DPH5 A0A182MGX3 A0A336K6M9 A0A182SCG8 A0A146LYJ7 N6U7R3 U4U011 A0A0P5S2K0 H3B0T0 A0A1D2MGZ7 A0A091R5P2 A0A2Y9STG5 A0A383YXL4 A0A2U3V077 A0A340Y961 A0A0F7YZC5 V8NUH3 A0A2Y9LPD4 A0A094L069 A0A2K5MCV3 A0A2R8ZFC6 H2QB64 A0A024R6U0 Q96DX4 A0A2J8VZ28 Q5R881 Q95LP3 G7Q173 G7NPT9 H0VUL8 Q28E18
E2A5D9 F4WU09 A0A151WWB8 A0A158P114 E2BTH6 A0A195DKG7 A0A026VU71 E9IE86 A0A2J7QJA0 A0A151JUR1 A0A0C9PSP3 A0A0L7QXQ5 A0A310S5F5 A0A232F4I1 A0A0M9A0T5 A0A154P927 A0A2W1BXX5 A0A1Y1M940 A0A2A3EHF8 K7IW25 A0A224Z6A8 A0A131YHF9 A0A131X8M0 L7M7R1 A0A1W4WNC8 E0W1N9 T1IHI8 A0A0P4W5Y0 A0A139WM94 A0A084W854 V5HK78 A0A1B6C6Q6 A0A2R5LBY4 A0A0P4XG14 A0A0P5BBS5 A0A0P5KAI0 A0A1Q3F9D4 A0A293LCM5 A0A182TSS6 A0A1Q3F995 A0A182W705 A0A182JGC5 E9HG27 A0A182R105 A0A1Q3F9D7 A0A182NUG3 A0A1S4GXE5 A0A182HUL7 A0A182L1D3 A0A182WU01 A0A182VHE8 Q17FP0 A0A182JPP5 B0WCT8 A0A182PPL0 W5J8V1 A0A182RCS6 A0A069DVB3 A0A182YQX9 A0A023F3S7 A0A0V0G6F2 A0A1B6I9K8 A0A1B6LFK8 A0A226DPH5 A0A182MGX3 A0A336K6M9 A0A182SCG8 A0A146LYJ7 N6U7R3 U4U011 A0A0P5S2K0 H3B0T0 A0A1D2MGZ7 A0A091R5P2 A0A2Y9STG5 A0A383YXL4 A0A2U3V077 A0A340Y961 A0A0F7YZC5 V8NUH3 A0A2Y9LPD4 A0A094L069 A0A2K5MCV3 A0A2R8ZFC6 H2QB64 A0A024R6U0 Q96DX4 A0A2J8VZ28 Q5R881 Q95LP3 G7Q173 G7NPT9 H0VUL8 Q28E18
PDB
5VSN
E-value=1.79559e-10,
Score=159
Ontologies
GO
Topology
Subcellular location
Secreted
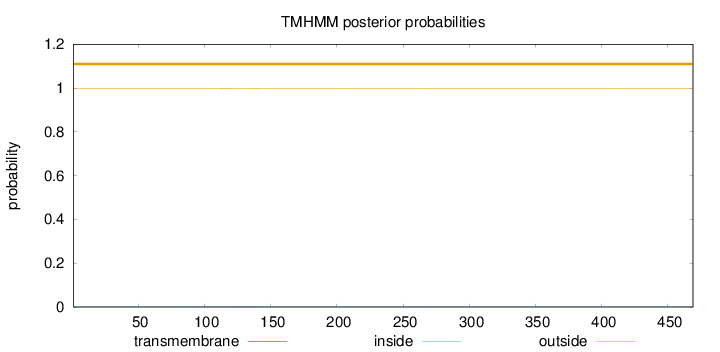
Length:
469
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.06613
Exp number, first 60 AAs:
0.00046
Total prob of N-in:
0.00326
outside
1 - 469
Population Genetic Test Statistics
Pi
27.133476
Theta
21.613301
Tajima's D
0.356133
CLR
0.077477
CSRT
0.474876256187191
Interpretation
Uncertain
