Gene
KWMTBOMO00425
Pre Gene Modal
BGIBMGA000488
Annotation
PREDICTED:_uncharacterized_protein_LOC106719909_[Papilio_machaon]
Full name
Protein grindelwald
Location in the cell
Nuclear Reliability : 2.561
Sequence
CDS
ATGTCGGGATTCGTGTGCGTCCTGATTCTCGCCGGCGTCGCCTCCGCTCAAATCACCCTGGACGGCATCAGATGCGGTCAGCTGATATGTCAGCTAGACGAATACTGCTCCCCGGAGACCAACCGATGCGCTCCGTGCAATGTTGTCTGCAATAAAACACACCATAACTACGATAGCGGCCTTTGCGTTAAGGAATGCCAAGGTTATCTATTGGATTTGAGGTACATGCGACGTTCGGAGACGGCCCCGCCACCCAACGATTTGGGAAGCATACACCGTGAAGCTCAAACTGCTCTGATCGTCAGCGGTGTGGCTCTGGCAGTACTGGTCCTAGTTCTCATCATAGTATGCAGAGGAAGAGTCTCTTGGAGATATTTAAAGCAAAAGTTCCAGCCAGCCAAGAACCGCGTGAAGCAGTACCCTAATGACTTGACGCATCACAACCCTCATGCCGAAGCGCCGAAACCGAAACCCGAACTCAAGCTCGAAATACGAAACCCGGATCCCCTAAGACGAACGAACCAACCTCTCAACGTGAAGGACTTAGAGACCAGAACCTCACAGTCGGATAAAAGTCAAGGCGCAACCACGCCAAAGACGATAAGCACCGCTCTTAGTAACAGGCATCCGGCTGAAGATACGACCTTAGATTTCTCTTATGATAATATGGGGATGAACGTGACTCCCCCCGGACAACCGGCTGCCAGTCACAAATTCTAA
Protein
MSGFVCVLILAGVASAQITLDGIRCGQLICQLDEYCSPETNRCAPCNVVCNKTHHNYDSGLCVKECQGYLLDLRYMRRSETAPPPNDLGSIHREAQTALIVSGVALAVLVLVLIIVCRGRVSWRYLKQKFQPAKNRVKQYPNDLTHHNPHAEAPKPKPELKLEIRNPDPLRRTNQPLNVKDLETRTSQSDKSQGATTPKTISTALSNRHPAEDTTLDFSYDNMGMNVTPPGQPAASHKF
Summary
Description
Acts as a receptor for egr. Plays a role in activation of JNK signaling and is required for egr-induced apoptosis. May also play an egr-independent role in cell proliferation.
Subunit
Interacts with egr, Traf6/TRAF2 and veli (via PDZ domain).
Miscellaneous
The name 'grindelwald' comes from the village at the foot of the Eiger mountain in Switzerland after which the egr gene is named.
Keywords
Cell membrane
Complete proteome
Glycoprotein
Membrane
Receptor
Reference proteome
Signal
Transmembrane
Transmembrane helix
Feature
chain Protein grindelwald
Uniprot
H9ITA8
A0A2H1WL56
A0A1E1W924
A0A194QN23
A0A212F9Q4
A0A194PGU2
+ More
A0A0L7L2F5 A0A1Y1LAD1 A0A1L8D9U4 A0A026X441 A0A0K8TP94 A0A139WI31 A0A158NS59 A0A195DE91 E9IR18 A0A195BCA1 A0A195FW85 N6UAB3 A0A088ARR9 E2A7P7 A0A1J1I659 F4WJQ3 A0A195BYX2 A0A151WH32 E2BD55 A0A1W4V9A8 A0A1W4WBK7 A0A1W4WBZ8 A0A1B6KZE1 A0A067QPA0 A0A1B6CQL4 A0A182NU10 A0A1B6EVB2 A0A182XFW7 A0A182VMS6 A0A2M3Z7J0 A0A2M3Z772 B4P9U6 A0A182UCP6 A0A182QBV2 A0A182L0H7 Q9VJ83 A0A0A9X5S2 A0A0J9R3Z9 A0A1S4GY44 A0A1W4VLD0 B4LTB8 A0A1W4WC23 B3NMB1 A0A182PLS7 A0A1W4VZX9 A0A1A9W0L6 A0A0Q9WBW7 A0A2M4DPE0 A0A182F3H1 A0A3B0K1Y2 A0A1W7R593 A0A2M4AB19 A0A2M4BZ85 A0A2M4BZB0 B4I5H2 A0A2M4AAL4 W5J948 B3MM20 A0A3B0JFL6 A0A2M4DPC5 A0A182K0V5 Q29MP9 Q17K49 A0A2S2NS13 W8B179 W8BZW9 A0A0P8Y325 A0A182J4K1 B4JDL8 A0A182RDS8 A0A182MTC3 A0A0R3NY52 A0A2R7VRS3 A0A0A1X6U9 A0A182YDT0 A0A1I8PEP6 A0A0P4VJ43 A0A0L0CCM1
A0A0L7L2F5 A0A1Y1LAD1 A0A1L8D9U4 A0A026X441 A0A0K8TP94 A0A139WI31 A0A158NS59 A0A195DE91 E9IR18 A0A195BCA1 A0A195FW85 N6UAB3 A0A088ARR9 E2A7P7 A0A1J1I659 F4WJQ3 A0A195BYX2 A0A151WH32 E2BD55 A0A1W4V9A8 A0A1W4WBK7 A0A1W4WBZ8 A0A1B6KZE1 A0A067QPA0 A0A1B6CQL4 A0A182NU10 A0A1B6EVB2 A0A182XFW7 A0A182VMS6 A0A2M3Z7J0 A0A2M3Z772 B4P9U6 A0A182UCP6 A0A182QBV2 A0A182L0H7 Q9VJ83 A0A0A9X5S2 A0A0J9R3Z9 A0A1S4GY44 A0A1W4VLD0 B4LTB8 A0A1W4WC23 B3NMB1 A0A182PLS7 A0A1W4VZX9 A0A1A9W0L6 A0A0Q9WBW7 A0A2M4DPE0 A0A182F3H1 A0A3B0K1Y2 A0A1W7R593 A0A2M4AB19 A0A2M4BZ85 A0A2M4BZB0 B4I5H2 A0A2M4AAL4 W5J948 B3MM20 A0A3B0JFL6 A0A2M4DPC5 A0A182K0V5 Q29MP9 Q17K49 A0A2S2NS13 W8B179 W8BZW9 A0A0P8Y325 A0A182J4K1 B4JDL8 A0A182RDS8 A0A182MTC3 A0A0R3NY52 A0A2R7VRS3 A0A0A1X6U9 A0A182YDT0 A0A1I8PEP6 A0A0P4VJ43 A0A0L0CCM1
Pubmed
19121390
26354079
22118469
26227816
28004739
24508170
+ More
30249741 26369729 18362917 19820115 21347285 21282665 23537049 20798317 21719571 24845553 17994087 17550304 20966253 10731132 12537572 25874673 25401762 26823975 22936249 12364791 20920257 23761445 15632085 17510324 24495485 25830018 25244985 27129103 26108605
30249741 26369729 18362917 19820115 21347285 21282665 23537049 20798317 21719571 24845553 17994087 17550304 20966253 10731132 12537572 25874673 25401762 26823975 22936249 12364791 20920257 23761445 15632085 17510324 24495485 25830018 25244985 27129103 26108605
EMBL
BABH01007283
BABH01007284
ODYU01009406
SOQ53811.1
GDQN01007581
JAT83473.1
+ More
KQ461196 KPJ06375.1 AGBW02009564 OWR50467.1 KQ459605 KPI91924.1 JTDY01003461 KOB69521.1 GEZM01061259 GEZM01061256 JAV70599.1 GFDF01010994 JAV03090.1 KK107031 QOIP01000008 EZA62129.1 RLU19284.1 GDAI01001396 JAI16207.1 KQ971342 KYB27633.1 ADTU01024524 KQ980989 KYN10744.1 GL765043 EFZ16966.1 KQ976528 KYM81829.1 KQ981208 KYN44686.1 APGK01032707 KB740848 ENN78665.1 GL437382 EFN70515.1 CVRI01000038 CRK94422.1 GL888186 EGI65545.1 KQ978457 KYM93849.1 KQ983136 KYQ47115.1 GL447569 EFN86363.1 GEBQ01031309 GEBQ01028440 GEBQ01023154 GEBQ01017119 JAT08668.1 JAT11537.1 JAT16823.1 JAT22858.1 KK853097 KDR11445.1 GEDC01021630 GEDC01007028 JAS15668.1 JAS30270.1 GECZ01030817 GECZ01027833 GECZ01003095 JAS38952.1 JAS41936.1 JAS66674.1 GGFM01003748 MBW24499.1 GGFM01003616 MBW24367.1 CM000158 EDW90287.2 AXCN02000809 AE014134 AY084146 AAL89884.1 GBHO01028260 GBRD01008136 GBRD01008134 GDHC01012356 GDHC01009407 GDHC01000299 JAG15344.1 JAG57685.1 JAQ06273.1 JAQ09222.1 JAQ18330.1 CM002910 KMY90818.1 AAAB01008964 CH940649 EDW64960.1 CH954179 EDV54782.2 KRF81954.1 GGFL01015246 MBW79424.1 OUUW01000004 SPP79011.1 GEHC01001298 JAV46347.1 GGFK01004629 MBW37950.1 GGFJ01009212 MBW58353.1 GGFJ01009211 MBW58352.1 CH480822 EDW55628.1 GGFK01004498 MBW37819.1 ADMH02001920 ETN60506.1 CH902620 EDV30835.1 SPP79012.1 GGFL01015245 MBW79423.1 CH379060 EAL33644.3 CH477228 EAT47044.1 GGMR01006927 MBY19546.1 GAMC01011710 GAMC01011708 JAB94845.1 GAMC01011711 GAMC01011709 JAB94844.1 KPU73075.1 CH916368 EDW03388.1 AXCM01001334 KRT04085.1 KK854020 PTY09531.1 GBXI01017335 GBXI01007475 GBXI01003425 JAC96956.1 JAD06817.1 JAD10867.1 GDKW01002462 JAI54133.1 JRES01000592 KNC30000.1
KQ461196 KPJ06375.1 AGBW02009564 OWR50467.1 KQ459605 KPI91924.1 JTDY01003461 KOB69521.1 GEZM01061259 GEZM01061256 JAV70599.1 GFDF01010994 JAV03090.1 KK107031 QOIP01000008 EZA62129.1 RLU19284.1 GDAI01001396 JAI16207.1 KQ971342 KYB27633.1 ADTU01024524 KQ980989 KYN10744.1 GL765043 EFZ16966.1 KQ976528 KYM81829.1 KQ981208 KYN44686.1 APGK01032707 KB740848 ENN78665.1 GL437382 EFN70515.1 CVRI01000038 CRK94422.1 GL888186 EGI65545.1 KQ978457 KYM93849.1 KQ983136 KYQ47115.1 GL447569 EFN86363.1 GEBQ01031309 GEBQ01028440 GEBQ01023154 GEBQ01017119 JAT08668.1 JAT11537.1 JAT16823.1 JAT22858.1 KK853097 KDR11445.1 GEDC01021630 GEDC01007028 JAS15668.1 JAS30270.1 GECZ01030817 GECZ01027833 GECZ01003095 JAS38952.1 JAS41936.1 JAS66674.1 GGFM01003748 MBW24499.1 GGFM01003616 MBW24367.1 CM000158 EDW90287.2 AXCN02000809 AE014134 AY084146 AAL89884.1 GBHO01028260 GBRD01008136 GBRD01008134 GDHC01012356 GDHC01009407 GDHC01000299 JAG15344.1 JAG57685.1 JAQ06273.1 JAQ09222.1 JAQ18330.1 CM002910 KMY90818.1 AAAB01008964 CH940649 EDW64960.1 CH954179 EDV54782.2 KRF81954.1 GGFL01015246 MBW79424.1 OUUW01000004 SPP79011.1 GEHC01001298 JAV46347.1 GGFK01004629 MBW37950.1 GGFJ01009212 MBW58353.1 GGFJ01009211 MBW58352.1 CH480822 EDW55628.1 GGFK01004498 MBW37819.1 ADMH02001920 ETN60506.1 CH902620 EDV30835.1 SPP79012.1 GGFL01015245 MBW79423.1 CH379060 EAL33644.3 CH477228 EAT47044.1 GGMR01006927 MBY19546.1 GAMC01011710 GAMC01011708 JAB94845.1 GAMC01011711 GAMC01011709 JAB94844.1 KPU73075.1 CH916368 EDW03388.1 AXCM01001334 KRT04085.1 KK854020 PTY09531.1 GBXI01017335 GBXI01007475 GBXI01003425 JAC96956.1 JAD06817.1 JAD10867.1 GDKW01002462 JAI54133.1 JRES01000592 KNC30000.1
Proteomes
UP000005204
UP000053240
UP000007151
UP000053268
UP000037510
UP000053097
+ More
UP000279307 UP000007266 UP000005205 UP000078492 UP000078540 UP000078541 UP000019118 UP000005203 UP000000311 UP000183832 UP000007755 UP000078542 UP000075809 UP000008237 UP000192221 UP000027135 UP000075884 UP000076407 UP000075903 UP000002282 UP000075902 UP000075886 UP000075882 UP000000803 UP000008792 UP000008711 UP000075885 UP000091820 UP000069272 UP000268350 UP000001292 UP000000673 UP000007801 UP000075881 UP000001819 UP000008820 UP000075880 UP000001070 UP000075900 UP000075883 UP000076408 UP000095300 UP000037069
UP000279307 UP000007266 UP000005205 UP000078492 UP000078540 UP000078541 UP000019118 UP000005203 UP000000311 UP000183832 UP000007755 UP000078542 UP000075809 UP000008237 UP000192221 UP000027135 UP000075884 UP000076407 UP000075903 UP000002282 UP000075902 UP000075886 UP000075882 UP000000803 UP000008792 UP000008711 UP000075885 UP000091820 UP000069272 UP000268350 UP000001292 UP000000673 UP000007801 UP000075881 UP000001819 UP000008820 UP000075880 UP000001070 UP000075900 UP000075883 UP000076408 UP000095300 UP000037069
ProteinModelPortal
H9ITA8
A0A2H1WL56
A0A1E1W924
A0A194QN23
A0A212F9Q4
A0A194PGU2
+ More
A0A0L7L2F5 A0A1Y1LAD1 A0A1L8D9U4 A0A026X441 A0A0K8TP94 A0A139WI31 A0A158NS59 A0A195DE91 E9IR18 A0A195BCA1 A0A195FW85 N6UAB3 A0A088ARR9 E2A7P7 A0A1J1I659 F4WJQ3 A0A195BYX2 A0A151WH32 E2BD55 A0A1W4V9A8 A0A1W4WBK7 A0A1W4WBZ8 A0A1B6KZE1 A0A067QPA0 A0A1B6CQL4 A0A182NU10 A0A1B6EVB2 A0A182XFW7 A0A182VMS6 A0A2M3Z7J0 A0A2M3Z772 B4P9U6 A0A182UCP6 A0A182QBV2 A0A182L0H7 Q9VJ83 A0A0A9X5S2 A0A0J9R3Z9 A0A1S4GY44 A0A1W4VLD0 B4LTB8 A0A1W4WC23 B3NMB1 A0A182PLS7 A0A1W4VZX9 A0A1A9W0L6 A0A0Q9WBW7 A0A2M4DPE0 A0A182F3H1 A0A3B0K1Y2 A0A1W7R593 A0A2M4AB19 A0A2M4BZ85 A0A2M4BZB0 B4I5H2 A0A2M4AAL4 W5J948 B3MM20 A0A3B0JFL6 A0A2M4DPC5 A0A182K0V5 Q29MP9 Q17K49 A0A2S2NS13 W8B179 W8BZW9 A0A0P8Y325 A0A182J4K1 B4JDL8 A0A182RDS8 A0A182MTC3 A0A0R3NY52 A0A2R7VRS3 A0A0A1X6U9 A0A182YDT0 A0A1I8PEP6 A0A0P4VJ43 A0A0L0CCM1
A0A0L7L2F5 A0A1Y1LAD1 A0A1L8D9U4 A0A026X441 A0A0K8TP94 A0A139WI31 A0A158NS59 A0A195DE91 E9IR18 A0A195BCA1 A0A195FW85 N6UAB3 A0A088ARR9 E2A7P7 A0A1J1I659 F4WJQ3 A0A195BYX2 A0A151WH32 E2BD55 A0A1W4V9A8 A0A1W4WBK7 A0A1W4WBZ8 A0A1B6KZE1 A0A067QPA0 A0A1B6CQL4 A0A182NU10 A0A1B6EVB2 A0A182XFW7 A0A182VMS6 A0A2M3Z7J0 A0A2M3Z772 B4P9U6 A0A182UCP6 A0A182QBV2 A0A182L0H7 Q9VJ83 A0A0A9X5S2 A0A0J9R3Z9 A0A1S4GY44 A0A1W4VLD0 B4LTB8 A0A1W4WC23 B3NMB1 A0A182PLS7 A0A1W4VZX9 A0A1A9W0L6 A0A0Q9WBW7 A0A2M4DPE0 A0A182F3H1 A0A3B0K1Y2 A0A1W7R593 A0A2M4AB19 A0A2M4BZ85 A0A2M4BZB0 B4I5H2 A0A2M4AAL4 W5J948 B3MM20 A0A3B0JFL6 A0A2M4DPC5 A0A182K0V5 Q29MP9 Q17K49 A0A2S2NS13 W8B179 W8BZW9 A0A0P8Y325 A0A182J4K1 B4JDL8 A0A182RDS8 A0A182MTC3 A0A0R3NY52 A0A2R7VRS3 A0A0A1X6U9 A0A182YDT0 A0A1I8PEP6 A0A0P4VJ43 A0A0L0CCM1
Ontologies
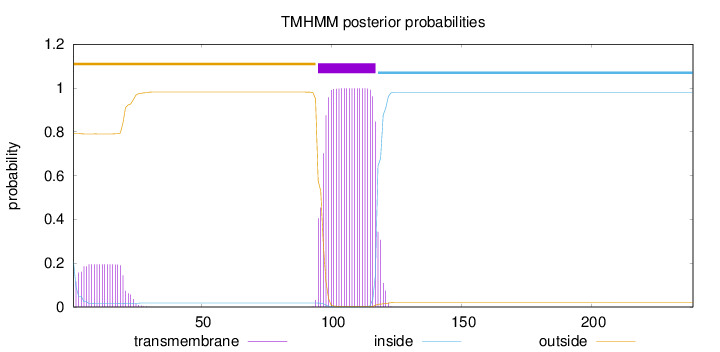
Topology
Subcellular location
Apical cell membrane
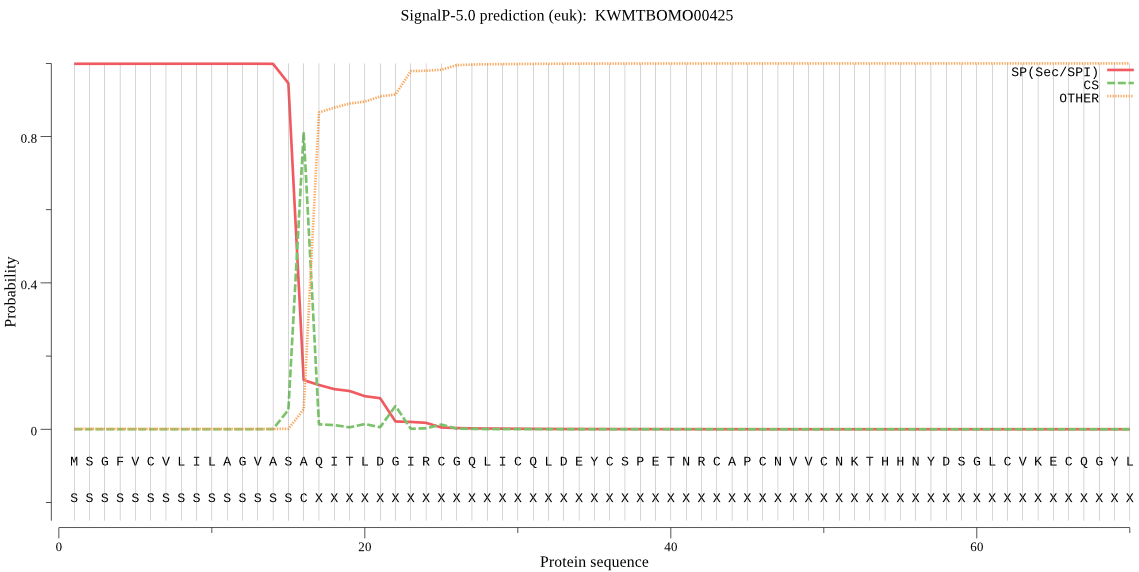
SignalP
Position: 1 - 16,
Likelihood: 0.999002
Length:
239
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
25.78395
Exp number, first 60 AAs:
3.73489
Total prob of N-in:
0.20617
outside
1 - 94
TMhelix
95 - 117
inside
118 - 239
Population Genetic Test Statistics
Pi
17.535452
Theta
21.052772
Tajima's D
-0.877304
CLR
0.052352
CSRT
0.15959202039898
Interpretation
Uncertain
