Gene
KWMTBOMO00410
Pre Gene Modal
BGIBMGA000732
Annotation
Cytochrome_b-c1_complex_subunit_Rieske?_mitochondrial_[Operophtera_brumata]
Full name
Cytochrome b-c1 complex subunit Rieske, mitochondrial
Alternative Name
Complex III subunit 5
Cytochrome b-c1 complex subunit 5
Rieske iron-sulfur protein
Rieske protein UQCRFS1
Ubiquinol-cytochrome c reductase iron-sulfur subunit
Cytochrome b-c1 complex subunit 5
Rieske iron-sulfur protein
Rieske protein UQCRFS1
Ubiquinol-cytochrome c reductase iron-sulfur subunit
Location in the cell
Nuclear Reliability : 0.963
Sequence
CDS
ATGAATGTTCTAAATAATGCGCGTGCAATTTGTCCTAATCTTTGGACCGGGCTGTGCTGTCAGAATTTAACTTGTATAAAAACTGTACGGGAGGTGCGACGAAAACGAGCGTCTGTCAATAAAAGCGATGCACCTCTAACGAGATTTTGTAGAAACAATCTTGAGCAAATACCGAGTCTGAGTATGTGGGTACAAGTGCGATACCACAAAGAATGTCCTCGTCTTCACCGCGATATCGAATTCCCAAACTTTAATCCGTACAGGAAAGATCAATACAAGGATGTGAGAAAGACAAGATGGGGTGCGGGCGACGAGAAGTCCGGAGTGACCTACACGATTGGCTTCTTCGGATTGCTCTGTGGATCGTATGCTGTAAAATCCGAGCTAGTACATTTCATTTCTTACATGGCCGCAGCCGCTGACGTGTTAGCTATGGCATCGATAGAAGTAGATCTGACCAAAATCACACCCGGGGCTTGTGTCTCATACAAATGGCGTGGAAAACCGCTTTTCATTAAGCGTAGAACTCAAGCAGACATCGATATTGAGGCAAAGACTCCATTGTCGGTTCTACGCGATCCAGAGACCCCGGAACAACGTACTACGATCCCGGAGTGGCTAATAGTGATTGGAATTTGTACTCATTTGGGATGTGTGCCGATCGCAAATGCTGGTGACTATCCGGGAGGTTTTTACTGTCCCTGCCACGGTAGTCATTACGATAACGTCGGTAGAGCTCGCAAAGGACCCGCGCCTTTGAATTTAGAGGTTCCCCCTTATAAGTTTGTTAGTGACTCTCTAGTAATTGTAGGATAA
Protein
MNVLNNARAICPNLWTGLCCQNLTCIKTVREVRRKRASVNKSDAPLTRFCRNNLEQIPSLSMWVQVRYHKECPRLHRDIEFPNFNPYRKDQYKDVRKTRWGAGDEKSGVTYTIGFFGLLCGSYAVKSELVHFISYMAAAADVLAMASIEVDLTKITPGACVSYKWRGKPLFIKRRTQADIDIEAKTPLSVLRDPETPEQRTTIPEWLIVIGICTHLGCVPIANAGDYPGGFYCPCHGSHYDNVGRARKGPAPLNLEVPPYKFVSDSLVIVG
Summary
Description
Component of the ubiquinol-cytochrome c reductase complex (complex III or cytochrome b-c1 complex), which is a respiratory chain that generates an electrochemical potential coupled to ATP synthesis.
Cytochrome b-c1 complex subunit Rieske, mitochondrial: Component of the mitochondrial ubiquinol-cytochrome c reductase complex dimer (complex III dimer), which is a respiratory chain that generates an electrochemical potential coupled to ATP synthesis (By similarity). Incorporation of UQCRFS1 is the penultimate step in complex III assembly (By similarity).
Cytochrome b-c1 complex subunit 9: Possible component of the mitochondrial ubiquinol-cytochrome c reductase complex dimer (complex III dimer), which is a respiratory chain that generates an electrochemical potential coupled to ATP synthesis (By similarity). UQCRFS1 undergoes proteolytic processing once it is incorporated in the complex III dimer, including this fragment, called subunit 9, which corresponds to the transit peptide (By similarity). The proteolytic processing is necessary for the correct insertion of UQCRFS1 in the complex III dimer, but the persistence of UQCRFS1-derived fragments may prevent newly imported UQCRFS1 to be processed and assembled into complex III and is detrimental for the complex III structure and function (By similarity). It is therefore unsure whether the UQCRFS1 fragments, including this fragment, are structural subunits (By similarity).
Cytochrome b-c1 complex subunit Rieske, mitochondrial: Component of the mitochondrial ubiquinol-cytochrome c reductase complex dimer (complex III dimer), which is a respiratory chain that generates an electrochemical potential coupled to ATP synthesis (By similarity). Incorporation of UQCRFS1 is the penultimate step in complex III assembly (By similarity).
Cytochrome b-c1 complex subunit 9: Possible component of the mitochondrial ubiquinol-cytochrome c reductase complex dimer (complex III dimer), which is a respiratory chain that generates an electrochemical potential coupled to ATP synthesis (By similarity). UQCRFS1 undergoes proteolytic processing once it is incorporated in the complex III dimer, including this fragment, called subunit 9, which corresponds to the transit peptide (By similarity). The proteolytic processing is necessary for the correct insertion of UQCRFS1 in the complex III dimer, but the persistence of UQCRFS1-derived fragments may prevent newly imported UQCRFS1 to be processed and assembled into complex III and is detrimental for the complex III structure and function (By similarity). It is therefore unsure whether the UQCRFS1 fragments, including this fragment, are structural subunits (By similarity).
Catalytic Activity
2 [Fe(III)cytochrome c](out) + a quinol = 2 [Fe(II)cytochrome c](out) + a quinone + 2 H(+)(out)
Cofactor
[2Fe-2S] cluster
Subunit
Binds to the mitochondrial respiratory complex III dimer. The monomeric module of the mitochondrial respiratory complex III contains 11 subunits: 3 respiratory subunits involved in its catalytic activity (cytochrome b, cytochrome c1 and Rieske protein UQCRFS1), 2 core proteins (UQCRC1/QCR1 and UQCRC2/QCR2) and 6 low-molecular weight proteins (UQCRH/QCR6, UQCRB/QCR7, UQCRQ/QCR8, UQCR10/QCR9, UQCR11/QCR10 and subunit 9, the cleavage product of Rieske protein UQCRFS1) (By similarity). Incorporation of the Rieske protein UQCRFS1 is the penultimate step in complex III assembly (By similarity). Interacts with TTC19, which is involved in the clearance of UQCRFS1 fragments (By similarity).
Miscellaneous
The Rieske protein is a high potential 2Fe-2S protein.
Keywords
2Fe-2S
Disulfide bond
Electron transport
Iron
Iron-sulfur
Membrane
Metal-binding
Mitochondrion
Mitochondrion inner membrane
Respiratory chain
Transit peptide
Translocase
Transmembrane
Transmembrane helix
Transport
3D-structure
Complete proteome
Reference proteome
Feature
chain Cytochrome b-c1 complex subunit 9
Uniprot
H9IU02
A0A0L7LQD2
A0A2A4JVB3
A0A2H1W0R7
A0A212F9N6
H9J0M9
+ More
B4JA98 B4KIH8 B3VTV1 A0A0P4Y3M5 A0A0P5IC96 A0A0P5EYS5 A0A0P4Z1V4 A0A162SVR7 A0A0P5NNX5 A0A0P5CVN3 A0A0P5Y6D5 A0A0P5L3B9 B4MW60 A0A0P5EY59 A0A0P5IT71 A0A0K8TIR8 A0A3M0K799 A0A0P5TK94 A0A212F5M9 A0A0P6E5P7 A0A0P4WSE7 B4LT56 A0A0P5BMS7 A0A0A9VZF5 A0A0P4ZNC7 A0A0P4WVP1 A0A0P4WVT4 A0A0P4ZSY3 Q69BJ9 A0A2K5D649 A0A0P4WPJ4 A0A0P5LF84 Q69BJ1 A0A0P5BW38 Q69BJ3 A0A0P5JPW9 A0A0P5D019 A0A0P5Z0W9 A0A0P5DZ82 A0A1L1RWY0 A0A0P5EDR0 A0A0P5EK99 A0A0N8CDX4 A0A0P5KR86 A0A099YV87 A0A0P5USJ2 A0A0P6IKA8 F1RNZ1 Q1G0W2 A0A091F3N1 Q5ZLR5 A0A093QH65 A0A1I8MQH0 A0A0P5APU4 A8JJ26 A0A0P5BEY8 R0JL39 A0A2P4T7P8 A0A1I8MQG2 A0A1Y1M8C3 U3KID7 T1PKU2 B3MK87 T1PDX2 A0A2K6UHY8 A0A0P5AD28 A0A1Q3FM73 K7GAR0 A0A091J6B5 A0A091U5Q4 Q69BJ8 A0A194PNL4 A0A1Q3FM50 A0A0N8AJI9 Q69BK7 Q69BJ7 A0A226PVD0 I4DKU3 H3A400 A0A0P6DGL8 A0A1I8PF44 K4FY92 A0A2K5TT07 A0A1I8PF34 A0A0M4EDJ2 S4PDC4 A0A0P4WSH9 B0XCK7 A0A287ASX6 Q69BJ5 A0A0A0MXF2 A0A091HVH6
B4JA98 B4KIH8 B3VTV1 A0A0P4Y3M5 A0A0P5IC96 A0A0P5EYS5 A0A0P4Z1V4 A0A162SVR7 A0A0P5NNX5 A0A0P5CVN3 A0A0P5Y6D5 A0A0P5L3B9 B4MW60 A0A0P5EY59 A0A0P5IT71 A0A0K8TIR8 A0A3M0K799 A0A0P5TK94 A0A212F5M9 A0A0P6E5P7 A0A0P4WSE7 B4LT56 A0A0P5BMS7 A0A0A9VZF5 A0A0P4ZNC7 A0A0P4WVP1 A0A0P4WVT4 A0A0P4ZSY3 Q69BJ9 A0A2K5D649 A0A0P4WPJ4 A0A0P5LF84 Q69BJ1 A0A0P5BW38 Q69BJ3 A0A0P5JPW9 A0A0P5D019 A0A0P5Z0W9 A0A0P5DZ82 A0A1L1RWY0 A0A0P5EDR0 A0A0P5EK99 A0A0N8CDX4 A0A0P5KR86 A0A099YV87 A0A0P5USJ2 A0A0P6IKA8 F1RNZ1 Q1G0W2 A0A091F3N1 Q5ZLR5 A0A093QH65 A0A1I8MQH0 A0A0P5APU4 A8JJ26 A0A0P5BEY8 R0JL39 A0A2P4T7P8 A0A1I8MQG2 A0A1Y1M8C3 U3KID7 T1PKU2 B3MK87 T1PDX2 A0A2K6UHY8 A0A0P5AD28 A0A1Q3FM73 K7GAR0 A0A091J6B5 A0A091U5Q4 Q69BJ8 A0A194PNL4 A0A1Q3FM50 A0A0N8AJI9 Q69BK7 Q69BJ7 A0A226PVD0 I4DKU3 H3A400 A0A0P6DGL8 A0A1I8PF44 K4FY92 A0A2K5TT07 A0A1I8PF34 A0A0M4EDJ2 S4PDC4 A0A0P4WSH9 B0XCK7 A0A287ASX6 Q69BJ5 A0A0A0MXF2 A0A091HVH6
EC Number
7.1.1.8
Pubmed
EMBL
BABH01007234
JTDY01000340
KOB77629.1
NWSH01000563
PCG75628.1
ODYU01005637
+ More
SOQ46685.1 AGBW02009564 OWR50455.1 BABH01010178 CH916368 EDW03772.1 CH933807 EDW11321.1 EU815629 HM210791 ACF21937.1 ADN06277.1 GDIP01236749 JAI86652.1 GDIQ01216818 JAK34907.1 GDIQ01268111 JAJ83613.1 GDIP01220602 JAJ02800.1 LRGB01000024 KZS21668.1 GDIQ01142342 JAL09384.1 GDIP01181098 JAJ42304.1 GDIP01062268 JAM41447.1 GDIQ01184529 JAK67196.1 CH963857 EDW75930.1 GDIQ01268321 JAJ83403.1 GDIQ01234064 JAK17661.1 GBRD01000816 JAG65005.1 QRBI01000116 RMC09053.1 GDIP01142610 JAL61104.1 AGBW02010163 OWR49045.1 GDIQ01067326 JAN27411.1 GDIP01252888 JAI70513.1 CH940649 EDW63887.1 GDIP01182795 JAJ40607.1 GBHO01044046 GDHC01010200 GDHC01004532 JAF99557.1 JAQ08429.1 JAQ14097.1 GDIP01211194 JAJ12208.1 GDIP01254486 JAI68915.1 GDIP01254422 JAI68979.1 GDIP01212151 JAJ11251.1 AY387510 AY387509 GDIP01254421 JAI68980.1 GDIQ01181165 JAK70560.1 AY387521 AAR32722.1 GDIP01179151 JAJ44251.1 AY387519 AAR32720.1 GDIQ01221465 JAK30260.1 GDIP01179150 JAJ44252.1 GDIP01050571 JAM53144.1 GDIP01149659 JAJ73743.1 GDIP01149660 JAJ73742.1 GDIP01147054 JAJ76348.1 GDIP01141255 JAL62459.1 GDIQ01187775 JAK63950.1 KL885851 KGL73296.1 GDIP01112302 JAL91412.1 GDIQ01003539 JAN91198.1 AEMK02000041 DQIR01031907 HCZ87382.1 DQ521661 ABF60225.1 KK719362 KFO63164.1 AJ719669 KL671666 KFW83422.1 GDIP01200699 JAJ22703.1 DS496302 EDO96122.1 GDIP01200700 JAJ22702.1 KB743565 EOA98015.1 PPHD01005941 POI32389.1 GEZM01037770 JAV81943.1 AGTO01021029 KA649334 KA649498 AFP63963.1 CH902620 EDV32471.1 KA646340 AFP60969.1 GDIP01200698 JAJ22704.1 GFDL01006459 JAV28586.1 AGCU01072991 AGCU01072992 KK501485 KFP15265.1 KK417835 KFQ85771.1 AY387512 AY387511 KQ459598 KPI94573.1 GFDL01006443 JAV28602.1 GDIQ01269547 JAJ82177.1 AY387494 AAR26321.1 AY387514 AY387513 AWGT02000007 OXB83991.1 AK401911 BAM18533.1 AFYH01216797 AFYH01216798 GDIQ01077917 JAN16820.1 JX052859 AFK11087.1 AQIA01035236 CP012523 ALC37967.1 GAIX01003721 JAA88839.1 GDIP01252187 JAI71214.1 DS232704 EDS44903.1 AY387517 AAR32718.1 AHZZ02014081 KL217815 KFO99147.1
SOQ46685.1 AGBW02009564 OWR50455.1 BABH01010178 CH916368 EDW03772.1 CH933807 EDW11321.1 EU815629 HM210791 ACF21937.1 ADN06277.1 GDIP01236749 JAI86652.1 GDIQ01216818 JAK34907.1 GDIQ01268111 JAJ83613.1 GDIP01220602 JAJ02800.1 LRGB01000024 KZS21668.1 GDIQ01142342 JAL09384.1 GDIP01181098 JAJ42304.1 GDIP01062268 JAM41447.1 GDIQ01184529 JAK67196.1 CH963857 EDW75930.1 GDIQ01268321 JAJ83403.1 GDIQ01234064 JAK17661.1 GBRD01000816 JAG65005.1 QRBI01000116 RMC09053.1 GDIP01142610 JAL61104.1 AGBW02010163 OWR49045.1 GDIQ01067326 JAN27411.1 GDIP01252888 JAI70513.1 CH940649 EDW63887.1 GDIP01182795 JAJ40607.1 GBHO01044046 GDHC01010200 GDHC01004532 JAF99557.1 JAQ08429.1 JAQ14097.1 GDIP01211194 JAJ12208.1 GDIP01254486 JAI68915.1 GDIP01254422 JAI68979.1 GDIP01212151 JAJ11251.1 AY387510 AY387509 GDIP01254421 JAI68980.1 GDIQ01181165 JAK70560.1 AY387521 AAR32722.1 GDIP01179151 JAJ44251.1 AY387519 AAR32720.1 GDIQ01221465 JAK30260.1 GDIP01179150 JAJ44252.1 GDIP01050571 JAM53144.1 GDIP01149659 JAJ73743.1 GDIP01149660 JAJ73742.1 GDIP01147054 JAJ76348.1 GDIP01141255 JAL62459.1 GDIQ01187775 JAK63950.1 KL885851 KGL73296.1 GDIP01112302 JAL91412.1 GDIQ01003539 JAN91198.1 AEMK02000041 DQIR01031907 HCZ87382.1 DQ521661 ABF60225.1 KK719362 KFO63164.1 AJ719669 KL671666 KFW83422.1 GDIP01200699 JAJ22703.1 DS496302 EDO96122.1 GDIP01200700 JAJ22702.1 KB743565 EOA98015.1 PPHD01005941 POI32389.1 GEZM01037770 JAV81943.1 AGTO01021029 KA649334 KA649498 AFP63963.1 CH902620 EDV32471.1 KA646340 AFP60969.1 GDIP01200698 JAJ22704.1 GFDL01006459 JAV28586.1 AGCU01072991 AGCU01072992 KK501485 KFP15265.1 KK417835 KFQ85771.1 AY387512 AY387511 KQ459598 KPI94573.1 GFDL01006443 JAV28602.1 GDIQ01269547 JAJ82177.1 AY387494 AAR26321.1 AY387514 AY387513 AWGT02000007 OXB83991.1 AK401911 BAM18533.1 AFYH01216797 AFYH01216798 GDIQ01077917 JAN16820.1 JX052859 AFK11087.1 AQIA01035236 CP012523 ALC37967.1 GAIX01003721 JAA88839.1 GDIP01252187 JAI71214.1 DS232704 EDS44903.1 AY387517 AAR32718.1 AHZZ02014081 KL217815 KFO99147.1
Proteomes
UP000005204
UP000037510
UP000218220
UP000007151
UP000001070
UP000009192
+ More
UP000076858 UP000007798 UP000269221 UP000008792 UP000233020 UP000053641 UP000008227 UP000052976 UP000000539 UP000053258 UP000095301 UP000016665 UP000007801 UP000233220 UP000007267 UP000053119 UP000053268 UP000198419 UP000008672 UP000095300 UP000233100 UP000092553 UP000002320 UP000028761 UP000054308
UP000076858 UP000007798 UP000269221 UP000008792 UP000233020 UP000053641 UP000008227 UP000052976 UP000000539 UP000053258 UP000095301 UP000016665 UP000007801 UP000233220 UP000007267 UP000053119 UP000053268 UP000198419 UP000008672 UP000095300 UP000233100 UP000092553 UP000002320 UP000028761 UP000054308
Interpro
Gene 3D
ProteinModelPortal
H9IU02
A0A0L7LQD2
A0A2A4JVB3
A0A2H1W0R7
A0A212F9N6
H9J0M9
+ More
B4JA98 B4KIH8 B3VTV1 A0A0P4Y3M5 A0A0P5IC96 A0A0P5EYS5 A0A0P4Z1V4 A0A162SVR7 A0A0P5NNX5 A0A0P5CVN3 A0A0P5Y6D5 A0A0P5L3B9 B4MW60 A0A0P5EY59 A0A0P5IT71 A0A0K8TIR8 A0A3M0K799 A0A0P5TK94 A0A212F5M9 A0A0P6E5P7 A0A0P4WSE7 B4LT56 A0A0P5BMS7 A0A0A9VZF5 A0A0P4ZNC7 A0A0P4WVP1 A0A0P4WVT4 A0A0P4ZSY3 Q69BJ9 A0A2K5D649 A0A0P4WPJ4 A0A0P5LF84 Q69BJ1 A0A0P5BW38 Q69BJ3 A0A0P5JPW9 A0A0P5D019 A0A0P5Z0W9 A0A0P5DZ82 A0A1L1RWY0 A0A0P5EDR0 A0A0P5EK99 A0A0N8CDX4 A0A0P5KR86 A0A099YV87 A0A0P5USJ2 A0A0P6IKA8 F1RNZ1 Q1G0W2 A0A091F3N1 Q5ZLR5 A0A093QH65 A0A1I8MQH0 A0A0P5APU4 A8JJ26 A0A0P5BEY8 R0JL39 A0A2P4T7P8 A0A1I8MQG2 A0A1Y1M8C3 U3KID7 T1PKU2 B3MK87 T1PDX2 A0A2K6UHY8 A0A0P5AD28 A0A1Q3FM73 K7GAR0 A0A091J6B5 A0A091U5Q4 Q69BJ8 A0A194PNL4 A0A1Q3FM50 A0A0N8AJI9 Q69BK7 Q69BJ7 A0A226PVD0 I4DKU3 H3A400 A0A0P6DGL8 A0A1I8PF44 K4FY92 A0A2K5TT07 A0A1I8PF34 A0A0M4EDJ2 S4PDC4 A0A0P4WSH9 B0XCK7 A0A287ASX6 Q69BJ5 A0A0A0MXF2 A0A091HVH6
B4JA98 B4KIH8 B3VTV1 A0A0P4Y3M5 A0A0P5IC96 A0A0P5EYS5 A0A0P4Z1V4 A0A162SVR7 A0A0P5NNX5 A0A0P5CVN3 A0A0P5Y6D5 A0A0P5L3B9 B4MW60 A0A0P5EY59 A0A0P5IT71 A0A0K8TIR8 A0A3M0K799 A0A0P5TK94 A0A212F5M9 A0A0P6E5P7 A0A0P4WSE7 B4LT56 A0A0P5BMS7 A0A0A9VZF5 A0A0P4ZNC7 A0A0P4WVP1 A0A0P4WVT4 A0A0P4ZSY3 Q69BJ9 A0A2K5D649 A0A0P4WPJ4 A0A0P5LF84 Q69BJ1 A0A0P5BW38 Q69BJ3 A0A0P5JPW9 A0A0P5D019 A0A0P5Z0W9 A0A0P5DZ82 A0A1L1RWY0 A0A0P5EDR0 A0A0P5EK99 A0A0N8CDX4 A0A0P5KR86 A0A099YV87 A0A0P5USJ2 A0A0P6IKA8 F1RNZ1 Q1G0W2 A0A091F3N1 Q5ZLR5 A0A093QH65 A0A1I8MQH0 A0A0P5APU4 A8JJ26 A0A0P5BEY8 R0JL39 A0A2P4T7P8 A0A1I8MQG2 A0A1Y1M8C3 U3KID7 T1PKU2 B3MK87 T1PDX2 A0A2K6UHY8 A0A0P5AD28 A0A1Q3FM73 K7GAR0 A0A091J6B5 A0A091U5Q4 Q69BJ8 A0A194PNL4 A0A1Q3FM50 A0A0N8AJI9 Q69BK7 Q69BJ7 A0A226PVD0 I4DKU3 H3A400 A0A0P6DGL8 A0A1I8PF44 K4FY92 A0A2K5TT07 A0A1I8PF34 A0A0M4EDJ2 S4PDC4 A0A0P4WSH9 B0XCK7 A0A287ASX6 Q69BJ5 A0A0A0MXF2 A0A091HVH6
PDB
5GUP
E-value=1.6579e-55,
Score=545
Ontologies
PATHWAY
GO
PANTHER
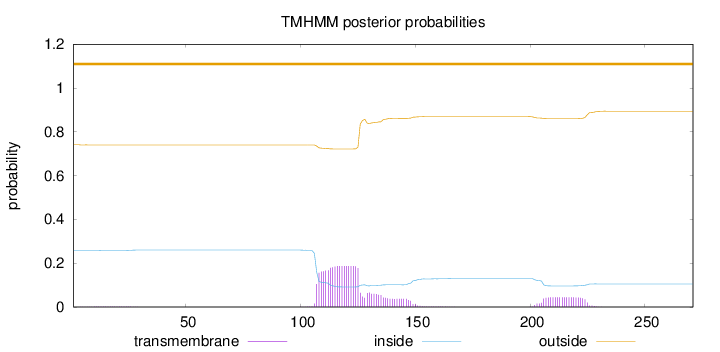
Topology
Subcellular location
Mitochondrion inner membrane
Length:
271
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.39942
Exp number, first 60 AAs:
0.03239
Total prob of N-in:
0.25874
outside
1 - 271
Population Genetic Test Statistics
Pi
19.951501
Theta
18.466431
Tajima's D
0.124116
CLR
0
CSRT
0.415979201039948
Interpretation
Uncertain
