Gene
KWMTBOMO00403
Pre Gene Modal
BGIBMGA000496
Annotation
PREDICTED:_peptide_transporter_family_1_isoform_X1_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.758
Sequence
CDS
ATGCGAACAATTTTGTCTTTGTACCTTCGAGACAAACTGGGCTTCACAGATAATGACGCCACAGTCATCTACCACGTGTTCACGATGTTTGCTTACTTCTTCCCGGTTCTCGGAGCCATGATTGCCGATGGATGGCTCGGAAGATTCAGGACCATATTTTATTTGTCACTGGTCTACGCTACGGGTAGTGTTCTAATTTCGCTGACCGCTATGCCCCCTATCGGCTTGCCGCAGTTTGAACTCACAATATTGGCGTTACTATTGATAGCTTTCGGAACGGGAGGTATTAAGCCCTGTGTGTCTGCTTTCGGAGGAGACCAATTCAAGCTACCAGAGCAGGCAAAATATCTCGGCTACTATTTTTCTCTCTTCTACTTCGCAATAAACGCGGGTAGTTTGATCTCCACCTTCTTGACTCCGATTCTGAGAGCTGACGTTCACTGTTTTGGAGACAGTGATTGCTTTTCATTGGCTTTCGGAGTCCCAGGGATTCTAATGGTGGTTTCGATTTTGATCTTCGTCGCTGGTCGAAGATTGTATGTAATCAAAAATCCATCAGGAAATGTTCTAGGGAAGGTGTCGACGTGTGTTGGGCATGCTGTGGTCAAATCCATCAAAAACAAACAGAAGAGAGACCACTGGTTGGACCACGCTGACGAGAAGTATGACAAGAGCCTAATCGAGGATATAAAGTCGCTGTTCAGGGTTTTGGTGCTATTCATTCCTCTACCAGTATTTTGGGCACTTTTTGATCAGCAGGGTTCCAGATGGACCTTCCAAGCTGACAGGATGGAACAAGACATAGGTAGCTGGACGCTTAAAGCTGATCAGATGCAAGTCCTCAATCCTTTCCTGATCTTAATATTTATTCCTCTCTTTGAAGTGGCAATTTATCCATTCCTAACTTGGTGCAGACTGATAAAGAAACCGCTACACAAAATGATTTGGGGTGGAATATTAGCAGCTTGCGCATTCCTTATTTCAGGAATAGTTGAATTAAATCTTTTGCCTACATACGGTACTCCGGTATCGGAAGGGTTGGCTCAACTCCGAGTGTACAACGGATTTGACTGTAATTTTACAATCAACACAGGAGATTTCAACGCCAACAATAACAATATTCATACTATTGGACCACTTTCGGTCTATGAGCATCTGGATATTAACGCAGATGGGTCTATTGAACTGCCGTATTATGTACGTGGCGAAGGAAATTGTTCGAATATCGTGTACAATGGAAATTTCTATTTAAAAGAAAATACTGCTAACTCTTTCTTCATTAATCGGGAGGGAATTTCTAACTTCACTGATAATAATGACAAAGCAATCAACGGAGTAAGCGTTAGGTTTTTGTCAAACGTCCAAACTTCGGTGACCGTTGCAATCGTGGAATCAAAGAAGAACAAAACTAAACTGTCACTGCAAAGCGATGATATCAGTCAGGTGAAGATCACCAAGGGCAATGGCGATGTTTTAGTGAACGACCAAAAAGTATTAGAAGGATATGCATTTAAATCTGGAGCAGTCTACACTATCAATGTTTACGAAGACACGGACGGTGCTTATCAAGCAAATCCAGTGATTATAACTCCACCAAATAGTATACACATTCTCTGGTTGATACCTCAGTACGTTGTGATGACTATGGGCGAAGTGATGTTTTCAGTAACTGGTTTAGAGTTCTCTTTCACACAAGCGCCAGCTAGTATGAAGTCTGTACTACAATCGGTCTGGCTTCTCACTGTCGCTTTTGGAAATTTGATCGTTGTTCTCATAGTGGAAGGGAACTTTTTGGATGCTCAGTGGAAAGAATTCTTCTTGTTCGCCGGTCTTATGTTATTGGACATGTTCATTTTCACGGCGATGGCGTTCAGATACAAGTACGTGGATCACAAATCGAGCACAGAGGAGGATATTGCCGTTGAAGAGATCAAAATGCCGGAAAAACCAACAATACAACAAAAAGACAATTAG
Protein
MRTILSLYLRDKLGFTDNDATVIYHVFTMFAYFFPVLGAMIADGWLGRFRTIFYLSLVYATGSVLISLTAMPPIGLPQFELTILALLLIAFGTGGIKPCVSAFGGDQFKLPEQAKYLGYYFSLFYFAINAGSLISTFLTPILRADVHCFGDSDCFSLAFGVPGILMVVSILIFVAGRRLYVIKNPSGNVLGKVSTCVGHAVVKSIKNKQKRDHWLDHADEKYDKSLIEDIKSLFRVLVLFIPLPVFWALFDQQGSRWTFQADRMEQDIGSWTLKADQMQVLNPFLILIFIPLFEVAIYPFLTWCRLIKKPLHKMIWGGILAACAFLISGIVELNLLPTYGTPVSEGLAQLRVYNGFDCNFTINTGDFNANNNNIHTIGPLSVYEHLDINADGSIELPYYVRGEGNCSNIVYNGNFYLKENTANSFFINREGISNFTDNNDKAINGVSVRFLSNVQTSVTVAIVESKKNKTKLSLQSDDISQVKITKGNGDVLVNDQKVLEGYAFKSGAVYTINVYEDTDGAYQANPVIITPPNSIHILWLIPQYVVMTMGEVMFSVTGLEFSFTQAPASMKSVLQSVWLLTVAFGNLIVVLIVEGNFLDAQWKEFFLFAGLMLLDMFIFTAMAFRYKYVDHKSSTEEDIAVEEIKMPEKPTIQQKDN
Summary
Similarity
Belongs to the PTR2/POT transporter (TC 2.A.17) family.
Uniprot
H9ITB6
A0A2A4JBI6
A0A212EGH0
S4P9F4
A0A194QN08
A0A2H1WTI7
+ More
A0A194PLF7 A0A2A4JCP3 A0A336KPU9 A0A1L8EGR4 A0A1I8MLE1 T1PEP1 A0A1Q3EVZ4 A0A1Q3EVN6 A0A182M5I4 U5EYN1 A0A182QYL0 A0A182MXD1 A0A034WFJ2 A0A0K8UNT4 A0A0A1WI31 A0A182R1W4 A0A182YC53 A0A182WD84 A0A1B0AMQ9 B0W557 A0A1A9YF24 A0A0K8VMD9 A0A1B0FDM9 K7J622 A0A084WM38 T1E1E0 A0A1I8NZD6 A0A1A9VWG9 A0A1A9Z1Z8 B4MB14 A0A1I8NZF8 A0A182KEV6 A0A182GHS3 A0A1S4G7L9 A0A2J7R2B8 A0A0C9RZK3 A0A2J7R2B4 A0A2J7R2B1 A0A182VMC2 A0A0C9PUC0 A0A182KT24 A0A0Q9WEG7 A0A1I8MLE4 A0A1A9WA00 B4NDF3 A0A182HPR3 A0A182X015 A0A182P1K6 A0A182TXA1 A0A0M3QYT3 A0A1L8E261 Q7PZY8 B4L741 W8C2V3 A0A0N8NZR0 B3MQY8 A0A0R1E9G3 A0A1L8E2G7 B4Q0H6 A0A232F6X3 B3NSS3 A0A1W4V1Z4 W8ANZ8 A0A1W4UPQ7 B4JX98 A0A182NVB3 Q8IRT1 W5JD76 B4R479 A0A195CBI2 B4I144 A0A0Q5T908 A0A3B0JDH5 A0A0A1WEF5 A0A3B0JC94 A0A0R3P0B0 A0A067RAG2 A0A182LRV8 X2JCG8 A0A1Q3FLX2 Q9W4P6 A0A182F432 D3PFE3 A0A182IYZ6 A0A182FVA2 B5DN49 A0A1B0CW25 A0A3B0JZL8 A0A0K8W9N9 A0A0K8U2K2 Q9U4G9 A0A084VBN9 A0A1A9VWG8
A0A194PLF7 A0A2A4JCP3 A0A336KPU9 A0A1L8EGR4 A0A1I8MLE1 T1PEP1 A0A1Q3EVZ4 A0A1Q3EVN6 A0A182M5I4 U5EYN1 A0A182QYL0 A0A182MXD1 A0A034WFJ2 A0A0K8UNT4 A0A0A1WI31 A0A182R1W4 A0A182YC53 A0A182WD84 A0A1B0AMQ9 B0W557 A0A1A9YF24 A0A0K8VMD9 A0A1B0FDM9 K7J622 A0A084WM38 T1E1E0 A0A1I8NZD6 A0A1A9VWG9 A0A1A9Z1Z8 B4MB14 A0A1I8NZF8 A0A182KEV6 A0A182GHS3 A0A1S4G7L9 A0A2J7R2B8 A0A0C9RZK3 A0A2J7R2B4 A0A2J7R2B1 A0A182VMC2 A0A0C9PUC0 A0A182KT24 A0A0Q9WEG7 A0A1I8MLE4 A0A1A9WA00 B4NDF3 A0A182HPR3 A0A182X015 A0A182P1K6 A0A182TXA1 A0A0M3QYT3 A0A1L8E261 Q7PZY8 B4L741 W8C2V3 A0A0N8NZR0 B3MQY8 A0A0R1E9G3 A0A1L8E2G7 B4Q0H6 A0A232F6X3 B3NSS3 A0A1W4V1Z4 W8ANZ8 A0A1W4UPQ7 B4JX98 A0A182NVB3 Q8IRT1 W5JD76 B4R479 A0A195CBI2 B4I144 A0A0Q5T908 A0A3B0JDH5 A0A0A1WEF5 A0A3B0JC94 A0A0R3P0B0 A0A067RAG2 A0A182LRV8 X2JCG8 A0A1Q3FLX2 Q9W4P6 A0A182F432 D3PFE3 A0A182IYZ6 A0A182FVA2 B5DN49 A0A1B0CW25 A0A3B0JZL8 A0A0K8W9N9 A0A0K8U2K2 Q9U4G9 A0A084VBN9 A0A1A9VWG8
Pubmed
19121390
22118469
23622113
26354079
25315136
25348373
+ More
25830018 25244985 20075255 24438588 24330624 17994087 26483478 17510324 20966253 12364791 24495485 17550304 28648823 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20920257 23761445 15632085 24845553 26109357 26109356
25830018 25244985 20075255 24438588 24330624 17994087 26483478 17510324 20966253 12364791 24495485 17550304 28648823 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 20920257 23761445 15632085 24845553 26109357 26109356
EMBL
BABH01007227
NWSH01002093
PCG69209.1
AGBW02015069
OWR40591.1
GAIX01005511
+ More
JAA87049.1 KQ461196 KPJ06360.1 ODYU01010950 SOQ56380.1 KQ459605 KPI91940.1 PCG69210.1 UFQS01000778 UFQT01000778 SSX06816.1 SSX27161.1 GFDG01001010 JAV17789.1 KA647206 AFP61835.1 GFDL01015667 JAV19378.1 GFDL01015675 JAV19370.1 AXCM01009176 AXCM01009177 GANO01001903 JAB57968.1 AXCN02001695 GAKP01004626 JAC54326.1 GDHF01023965 JAI28349.1 GBXI01015755 JAC98536.1 JXJN01000552 DS231841 EDS34804.1 GDHF01012262 JAI40052.1 CCAG010017640 ATLV01024373 KE525351 KFB51282.1 GALA01001764 JAA93088.1 CH940655 EDW66423.2 JXUM01064548 JXUM01064549 JXUM01064550 KQ562303 KXJ76196.1 NEVH01007841 PNF34979.1 GBYB01013556 JAG83323.1 PNF34978.1 PNF34977.1 GBYB01004958 JAG74725.1 KRF82674.1 CH964239 EDW82859.2 APCN01003267 CP012528 ALC48233.1 GFDF01001264 JAV12820.1 AAAB01008986 EAA00193.4 CH933812 EDW06187.1 GAMC01010461 JAB96094.1 CH902622 KPU75020.1 EDV34193.1 CM000162 KRK06028.1 GFDF01001265 JAV12819.1 EDX01260.1 KRK06027.1 NNAY01000849 OXU26213.1 CH954180 EDV45753.1 KQS29696.1 GAMC01020327 GAMC01020325 JAB86228.1 CH916376 EDV95374.1 AE014298 AAN09112.2 ADMH02001720 ETN61308.1 CM000366 EDX17021.1 KQ978023 KYM98055.1 CH480819 EDW53225.1 KQS29697.1 OUUW01000003 SPP78182.1 GBXI01017457 JAC96834.1 SPP78183.1 CH379064 KRT06727.1 KK852588 KDR20782.1 AXCM01000199 AHN59316.1 GFDL01006436 JAV28609.1 BT056251 AAF45901.2 ACL68698.1 BT120302 ADC53519.1 EDY72856.1 AJWK01031590 SPP78181.1 GDHF01004575 JAI47739.1 GDHF01031581 JAI20733.1 AF181635 AAD55421.1 ATLV01008553 KE524477 KFB35383.1
JAA87049.1 KQ461196 KPJ06360.1 ODYU01010950 SOQ56380.1 KQ459605 KPI91940.1 PCG69210.1 UFQS01000778 UFQT01000778 SSX06816.1 SSX27161.1 GFDG01001010 JAV17789.1 KA647206 AFP61835.1 GFDL01015667 JAV19378.1 GFDL01015675 JAV19370.1 AXCM01009176 AXCM01009177 GANO01001903 JAB57968.1 AXCN02001695 GAKP01004626 JAC54326.1 GDHF01023965 JAI28349.1 GBXI01015755 JAC98536.1 JXJN01000552 DS231841 EDS34804.1 GDHF01012262 JAI40052.1 CCAG010017640 ATLV01024373 KE525351 KFB51282.1 GALA01001764 JAA93088.1 CH940655 EDW66423.2 JXUM01064548 JXUM01064549 JXUM01064550 KQ562303 KXJ76196.1 NEVH01007841 PNF34979.1 GBYB01013556 JAG83323.1 PNF34978.1 PNF34977.1 GBYB01004958 JAG74725.1 KRF82674.1 CH964239 EDW82859.2 APCN01003267 CP012528 ALC48233.1 GFDF01001264 JAV12820.1 AAAB01008986 EAA00193.4 CH933812 EDW06187.1 GAMC01010461 JAB96094.1 CH902622 KPU75020.1 EDV34193.1 CM000162 KRK06028.1 GFDF01001265 JAV12819.1 EDX01260.1 KRK06027.1 NNAY01000849 OXU26213.1 CH954180 EDV45753.1 KQS29696.1 GAMC01020327 GAMC01020325 JAB86228.1 CH916376 EDV95374.1 AE014298 AAN09112.2 ADMH02001720 ETN61308.1 CM000366 EDX17021.1 KQ978023 KYM98055.1 CH480819 EDW53225.1 KQS29697.1 OUUW01000003 SPP78182.1 GBXI01017457 JAC96834.1 SPP78183.1 CH379064 KRT06727.1 KK852588 KDR20782.1 AXCM01000199 AHN59316.1 GFDL01006436 JAV28609.1 BT056251 AAF45901.2 ACL68698.1 BT120302 ADC53519.1 EDY72856.1 AJWK01031590 SPP78181.1 GDHF01004575 JAI47739.1 GDHF01031581 JAI20733.1 AF181635 AAD55421.1 ATLV01008553 KE524477 KFB35383.1
Proteomes
UP000005204
UP000218220
UP000007151
UP000053240
UP000053268
UP000095301
+ More
UP000075883 UP000075886 UP000075884 UP000075900 UP000076408 UP000075920 UP000092460 UP000002320 UP000092443 UP000092444 UP000002358 UP000030765 UP000095300 UP000078200 UP000092445 UP000008792 UP000075881 UP000069940 UP000249989 UP000235965 UP000075903 UP000075882 UP000091820 UP000007798 UP000075840 UP000076407 UP000075885 UP000075902 UP000092553 UP000007062 UP000009192 UP000007801 UP000002282 UP000215335 UP000008711 UP000192221 UP000001070 UP000000803 UP000000673 UP000000304 UP000078542 UP000001292 UP000268350 UP000001819 UP000027135 UP000069272 UP000075880 UP000092461
UP000075883 UP000075886 UP000075884 UP000075900 UP000076408 UP000075920 UP000092460 UP000002320 UP000092443 UP000092444 UP000002358 UP000030765 UP000095300 UP000078200 UP000092445 UP000008792 UP000075881 UP000069940 UP000249989 UP000235965 UP000075903 UP000075882 UP000091820 UP000007798 UP000075840 UP000076407 UP000075885 UP000075902 UP000092553 UP000007062 UP000009192 UP000007801 UP000002282 UP000215335 UP000008711 UP000192221 UP000001070 UP000000803 UP000000673 UP000000304 UP000078542 UP000001292 UP000268350 UP000001819 UP000027135 UP000069272 UP000075880 UP000092461
Pfam
PF00854 PTR2
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
H9ITB6
A0A2A4JBI6
A0A212EGH0
S4P9F4
A0A194QN08
A0A2H1WTI7
+ More
A0A194PLF7 A0A2A4JCP3 A0A336KPU9 A0A1L8EGR4 A0A1I8MLE1 T1PEP1 A0A1Q3EVZ4 A0A1Q3EVN6 A0A182M5I4 U5EYN1 A0A182QYL0 A0A182MXD1 A0A034WFJ2 A0A0K8UNT4 A0A0A1WI31 A0A182R1W4 A0A182YC53 A0A182WD84 A0A1B0AMQ9 B0W557 A0A1A9YF24 A0A0K8VMD9 A0A1B0FDM9 K7J622 A0A084WM38 T1E1E0 A0A1I8NZD6 A0A1A9VWG9 A0A1A9Z1Z8 B4MB14 A0A1I8NZF8 A0A182KEV6 A0A182GHS3 A0A1S4G7L9 A0A2J7R2B8 A0A0C9RZK3 A0A2J7R2B4 A0A2J7R2B1 A0A182VMC2 A0A0C9PUC0 A0A182KT24 A0A0Q9WEG7 A0A1I8MLE4 A0A1A9WA00 B4NDF3 A0A182HPR3 A0A182X015 A0A182P1K6 A0A182TXA1 A0A0M3QYT3 A0A1L8E261 Q7PZY8 B4L741 W8C2V3 A0A0N8NZR0 B3MQY8 A0A0R1E9G3 A0A1L8E2G7 B4Q0H6 A0A232F6X3 B3NSS3 A0A1W4V1Z4 W8ANZ8 A0A1W4UPQ7 B4JX98 A0A182NVB3 Q8IRT1 W5JD76 B4R479 A0A195CBI2 B4I144 A0A0Q5T908 A0A3B0JDH5 A0A0A1WEF5 A0A3B0JC94 A0A0R3P0B0 A0A067RAG2 A0A182LRV8 X2JCG8 A0A1Q3FLX2 Q9W4P6 A0A182F432 D3PFE3 A0A182IYZ6 A0A182FVA2 B5DN49 A0A1B0CW25 A0A3B0JZL8 A0A0K8W9N9 A0A0K8U2K2 Q9U4G9 A0A084VBN9 A0A1A9VWG8
A0A194PLF7 A0A2A4JCP3 A0A336KPU9 A0A1L8EGR4 A0A1I8MLE1 T1PEP1 A0A1Q3EVZ4 A0A1Q3EVN6 A0A182M5I4 U5EYN1 A0A182QYL0 A0A182MXD1 A0A034WFJ2 A0A0K8UNT4 A0A0A1WI31 A0A182R1W4 A0A182YC53 A0A182WD84 A0A1B0AMQ9 B0W557 A0A1A9YF24 A0A0K8VMD9 A0A1B0FDM9 K7J622 A0A084WM38 T1E1E0 A0A1I8NZD6 A0A1A9VWG9 A0A1A9Z1Z8 B4MB14 A0A1I8NZF8 A0A182KEV6 A0A182GHS3 A0A1S4G7L9 A0A2J7R2B8 A0A0C9RZK3 A0A2J7R2B4 A0A2J7R2B1 A0A182VMC2 A0A0C9PUC0 A0A182KT24 A0A0Q9WEG7 A0A1I8MLE4 A0A1A9WA00 B4NDF3 A0A182HPR3 A0A182X015 A0A182P1K6 A0A182TXA1 A0A0M3QYT3 A0A1L8E261 Q7PZY8 B4L741 W8C2V3 A0A0N8NZR0 B3MQY8 A0A0R1E9G3 A0A1L8E2G7 B4Q0H6 A0A232F6X3 B3NSS3 A0A1W4V1Z4 W8ANZ8 A0A1W4UPQ7 B4JX98 A0A182NVB3 Q8IRT1 W5JD76 B4R479 A0A195CBI2 B4I144 A0A0Q5T908 A0A3B0JDH5 A0A0A1WEF5 A0A3B0JC94 A0A0R3P0B0 A0A067RAG2 A0A182LRV8 X2JCG8 A0A1Q3FLX2 Q9W4P6 A0A182F432 D3PFE3 A0A182IYZ6 A0A182FVA2 B5DN49 A0A1B0CW25 A0A3B0JZL8 A0A0K8W9N9 A0A0K8U2K2 Q9U4G9 A0A084VBN9 A0A1A9VWG8
PDB
4UVM
E-value=3.15885e-31,
Score=339
Ontologies
GO
PANTHER
Topology
Subcellular location
Membrane
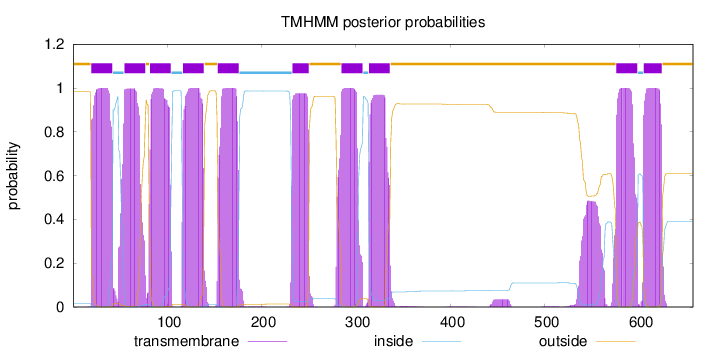
Length:
657
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
226.54901
Exp number, first 60 AAs:
31.35775
Total prob of N-in:
0.01591
POSSIBLE N-term signal
sequence
outside
1 - 19
TMhelix
20 - 42
inside
43 - 54
TMhelix
55 - 77
outside
78 - 81
TMhelix
82 - 104
inside
105 - 116
TMhelix
117 - 139
outside
140 - 153
TMhelix
154 - 176
inside
177 - 232
TMhelix
233 - 250
outside
251 - 284
TMhelix
285 - 307
inside
308 - 313
TMhelix
314 - 336
outside
337 - 575
TMhelix
576 - 598
inside
599 - 604
TMhelix
605 - 624
outside
625 - 657
Population Genetic Test Statistics
Pi
3.059995
Theta
15.206857
Tajima's D
-1.562502
CLR
0.070149
CSRT
0.0487475626218689
Interpretation
Uncertain
