Gene
KWMTBOMO00399
Annotation
PREDICTED:_LOW_QUALITY_PROTEIN:_circadian_locomoter_output_cycles_protein_kaput_[Bombyx_mori]
Transcription factor
Location in the cell
Nuclear Reliability : 2.898
Sequence
CDS
ATGGAAGACGACGGAGACGACAAAGATGATACTAAGAGGAGGACTCGTAACTTGAGTGAAAAAAAGCGACGGGATCAGTTTAATATGCTGGTCAATGAGCTTAGTTCAATGGTCTCAACGAACAACAGGAAGATGGATAAATCTACAGTACTCAAGTCTACAATATCCTTCCTGAAAAACCATAATGAAATAACAGTAAGATCTCGAGCACATGACATACAAGAAGATTGGAAACCAACTTTCCTATCAAATGAAGAGTTTACATACTTGGTTCTTGAAGCGTTAGAAGGATTTGTCATGGTTTTTTCTGCGTCAGGTCGAATCTACTACGTCTCCGAAAGCATTAAATCTCTTCTTGGTTACAATCCAACTGAAATTCTCAATAAGAGCATATTTGATTTGACGTTTGAAGAGGATCGCCCTAATTTATATAACATTCTACAGAACACAAATACCCCGAATCCCAAAGGTAAGTTTTCATTCAACCTCACTATTTAA
Protein
MEDDGDDKDDTKRRTRNLSEKKRRDQFNMLVNELSSMVSTNNRKMDKSTVLKSTISFLKNHNEITVRSRAHDIQEDWKPTFLSNEEFTYLVLEALEGFVMVFSASGRIYYVSESIKSLLGYNPTEILNKSIFDLTFEEDRPNLYNILQNTNTPNPKGKFSFNLTI
Summary
Uniprot
A0A2H1VR20
A0A1E1WCW9
A0A2W1BNQ5
G0YQM3
A0A1V0EMB5
A0A212EGJ4
+ More
A0A0N7BAL6 Q3ZTR5 A0A1E1WUU5 A0A2A4IWA8 A0A194PGW1 A0A194QLE2 Q6VRU6 A0A2J7QFH5 E0D6T2 L8B0S6 A0A1B0GJJ1 E2C765 A0A151WTM8 A0A336KII5 A0A068Q7V1 A0A310SLY2 A0A0L7RH10 A0A195BTA5 A0A0H4BGL7 A0A195FJ10 F4WUR0 L7Y4E9 A0A0M9A7L2 A0A195DM36 A0A158P2D5 A0A195D379 A0A2A3EIL5 A0A088A9H3 A0A3L8E090 A0A1B0D5T7 A0A1L8DH51 A0A1L8DGZ4 A0A154PGQ5 E2AY34 B0W3Y0 A0A0J7L871 A0A1L5YQJ2 A0A232EFE1 E0VT39 A0A182L2Y8 A0A182U1Q0 A0A182FCB7 H7CI07 U5EHU6 A0A182WKT4 A0A1B6CYF0 A0A1S4FWV2 Q16LQ2 A9XCF0 A0A2M3ZFQ8 N6UMH2 U4U4S1 A0A182PMI9 A0A182XMS6 A0A182Q7X5 A0A182VHW3 A0A182LWI6 A0A0F7DCF2 K7JC66 D6WIQ8 A0A182HWN3 Q7Q6R8 A0A182NIR3 A0A182YD49 A0A1Y1M421 A0A182RZS8 A0A084WA50 A0A182KDN3 A0A182IKV4 R4WQ03 A0A1J1I6S5 A0A2M4BB90 A0A2M4A2Y8 A0A2M4BB67 W5JIJ2 A0A1Q3G4C9 A0A1Q3G4C5 A0A2M3YYP5 A0A2M4A4J9 T1HDJ0 A0A1Q3G490 A0A1Q3G4K0 A0A023FAB5 A0A182H8U9 A0A146LMC7 A0A0K8SX26 A0A0A9W299 A0A1B0YYT9 E9IE69 A0A2S0X9U7 A0A0A1XCF3 Q2KPA5 D2Z1J8
A0A0N7BAL6 Q3ZTR5 A0A1E1WUU5 A0A2A4IWA8 A0A194PGW1 A0A194QLE2 Q6VRU6 A0A2J7QFH5 E0D6T2 L8B0S6 A0A1B0GJJ1 E2C765 A0A151WTM8 A0A336KII5 A0A068Q7V1 A0A310SLY2 A0A0L7RH10 A0A195BTA5 A0A0H4BGL7 A0A195FJ10 F4WUR0 L7Y4E9 A0A0M9A7L2 A0A195DM36 A0A158P2D5 A0A195D379 A0A2A3EIL5 A0A088A9H3 A0A3L8E090 A0A1B0D5T7 A0A1L8DH51 A0A1L8DGZ4 A0A154PGQ5 E2AY34 B0W3Y0 A0A0J7L871 A0A1L5YQJ2 A0A232EFE1 E0VT39 A0A182L2Y8 A0A182U1Q0 A0A182FCB7 H7CI07 U5EHU6 A0A182WKT4 A0A1B6CYF0 A0A1S4FWV2 Q16LQ2 A9XCF0 A0A2M3ZFQ8 N6UMH2 U4U4S1 A0A182PMI9 A0A182XMS6 A0A182Q7X5 A0A182VHW3 A0A182LWI6 A0A0F7DCF2 K7JC66 D6WIQ8 A0A182HWN3 Q7Q6R8 A0A182NIR3 A0A182YD49 A0A1Y1M421 A0A182RZS8 A0A084WA50 A0A182KDN3 A0A182IKV4 R4WQ03 A0A1J1I6S5 A0A2M4BB90 A0A2M4A2Y8 A0A2M4BB67 W5JIJ2 A0A1Q3G4C9 A0A1Q3G4C5 A0A2M3YYP5 A0A2M4A4J9 T1HDJ0 A0A1Q3G490 A0A1Q3G4K0 A0A023FAB5 A0A182H8U9 A0A146LMC7 A0A0K8SX26 A0A0A9W299 A0A1B0YYT9 E9IE69 A0A2S0X9U7 A0A0A1XCF3 Q2KPA5 D2Z1J8
Pubmed
28756777
22118469
16332522
26354079
12869551
20416313
+ More
23223372 20798317 21719571 23152747 21347285 30249741 28648823 20566863 20966253 17510324 23537049 20075255 18362917 19820115 12364791 14747013 17210077 25244985 28004739 24438588 23691247 20920257 23761445 25474469 26483478 26823975 25401762 27341138 21282665 29655930 25830018 16271708
23223372 20798317 21719571 23152747 21347285 30249741 28648823 20566863 20966253 17510324 23537049 20075255 18362917 19820115 12364791 14747013 17210077 25244985 28004739 24438588 23691247 20920257 23761445 25474469 26483478 26823975 25401762 27341138 21282665 29655930 25830018 16271708
EMBL
ODYU01003912
SOQ43260.1
GDQN01006212
JAT84842.1
KZ149977
PZC75911.1
+ More
HQ731032 AEJ38222.1 KY446807 ARB18436.1 AGBW02015069 OWR40606.1 KM233158 AKC42319.1 AY364477 AAR13011.1 GDQN01000264 JAT90790.1 NWSH01006407 PCG63462.1 KQ459605 KPI91944.1 KQ461196 KPJ06357.1 AY330486 AAR14936.1 NEVH01015298 PNF27334.1 AB550828 BAJ16353.1 AB738083 BAM76759.1 AJWK01021846 GL453340 EFN76178.1 KQ982753 KYQ51193.1 UFQS01000473 UFQT01000473 SSX04269.1 SSX24634.1 AB894249 BAP15889.1 KQ759885 OAD62154.1 KQ414594 KOC70150.1 KQ976408 KYM91193.1 KR706373 AKN63486.1 KQ981523 KYN40252.1 GL888375 EGI62057.1 JX948388 AGD94516.1 KQ435730 KOX77563.1 KQ980734 KYN13948.1 ADTU01007159 KQ976885 KYN07352.1 KZ288254 PBC30841.1 QOIP01000002 RLU25795.1 AJVK01000345 AJVK01000346 GFDF01008394 JAV05690.1 GFDF01008393 JAV05691.1 KQ434900 KZC11045.1 GL443762 EFN61630.1 DS231833 EDS32277.1 LBMM01000329 KMR01088.1 KX602314 APP94023.1 NNAY01005041 OXU17076.1 DS235759 EEB16545.1 AB704408 BAL72807.1 GANO01002879 JAB56992.1 GEDC01018854 JAS18444.1 CH477897 EAT35264.1 EF174303 ABO86537.1 GGFM01006509 MBW27260.1 APGK01026653 KB740635 KB631539 ENN79917.1 ERL84364.1 KB632107 ERL88874.1 AXCN02000420 AXCM01004081 KP147912 AKG92749.1 AAZX01001489 KQ971321 EFA01240.2 APCN01001511 AAAB01008960 EAA11642.4 GEZM01041677 JAV80231.1 ATLV01022043 KE525327 KFB47094.1 AK417766 BAN20981.1 CVRI01000043 CRK95960.1 GGFJ01001156 MBW50297.1 GGFK01001852 MBW35173.1 GGFJ01001142 MBW50283.1 ADMH02001414 ETN62614.1 GFDL01000388 JAV34657.1 GFDL01000383 JAV34662.1 GGFM01000629 MBW21380.1 GGFK01002247 MBW35568.1 ACPB03010898 GFDL01000420 JAV34625.1 GFDL01000315 JAV34730.1 GBBI01000290 JAC18422.1 JXUM01029554 KQ560856 KXJ80615.1 GDHC01009551 JAQ09078.1 GBRD01008110 JAG57711.1 GBHO01044614 JAF98989.1 KX014718 ANO53967.1 GL762558 EFZ21134.1 MG280775 AWC08577.1 GBXI01005665 JAD08627.1 AY842303 AAX44045.1 AB543754 BAI67740.1
HQ731032 AEJ38222.1 KY446807 ARB18436.1 AGBW02015069 OWR40606.1 KM233158 AKC42319.1 AY364477 AAR13011.1 GDQN01000264 JAT90790.1 NWSH01006407 PCG63462.1 KQ459605 KPI91944.1 KQ461196 KPJ06357.1 AY330486 AAR14936.1 NEVH01015298 PNF27334.1 AB550828 BAJ16353.1 AB738083 BAM76759.1 AJWK01021846 GL453340 EFN76178.1 KQ982753 KYQ51193.1 UFQS01000473 UFQT01000473 SSX04269.1 SSX24634.1 AB894249 BAP15889.1 KQ759885 OAD62154.1 KQ414594 KOC70150.1 KQ976408 KYM91193.1 KR706373 AKN63486.1 KQ981523 KYN40252.1 GL888375 EGI62057.1 JX948388 AGD94516.1 KQ435730 KOX77563.1 KQ980734 KYN13948.1 ADTU01007159 KQ976885 KYN07352.1 KZ288254 PBC30841.1 QOIP01000002 RLU25795.1 AJVK01000345 AJVK01000346 GFDF01008394 JAV05690.1 GFDF01008393 JAV05691.1 KQ434900 KZC11045.1 GL443762 EFN61630.1 DS231833 EDS32277.1 LBMM01000329 KMR01088.1 KX602314 APP94023.1 NNAY01005041 OXU17076.1 DS235759 EEB16545.1 AB704408 BAL72807.1 GANO01002879 JAB56992.1 GEDC01018854 JAS18444.1 CH477897 EAT35264.1 EF174303 ABO86537.1 GGFM01006509 MBW27260.1 APGK01026653 KB740635 KB631539 ENN79917.1 ERL84364.1 KB632107 ERL88874.1 AXCN02000420 AXCM01004081 KP147912 AKG92749.1 AAZX01001489 KQ971321 EFA01240.2 APCN01001511 AAAB01008960 EAA11642.4 GEZM01041677 JAV80231.1 ATLV01022043 KE525327 KFB47094.1 AK417766 BAN20981.1 CVRI01000043 CRK95960.1 GGFJ01001156 MBW50297.1 GGFK01001852 MBW35173.1 GGFJ01001142 MBW50283.1 ADMH02001414 ETN62614.1 GFDL01000388 JAV34657.1 GFDL01000383 JAV34662.1 GGFM01000629 MBW21380.1 GGFK01002247 MBW35568.1 ACPB03010898 GFDL01000420 JAV34625.1 GFDL01000315 JAV34730.1 GBBI01000290 JAC18422.1 JXUM01029554 KQ560856 KXJ80615.1 GDHC01009551 JAQ09078.1 GBRD01008110 JAG57711.1 GBHO01044614 JAF98989.1 KX014718 ANO53967.1 GL762558 EFZ21134.1 MG280775 AWC08577.1 GBXI01005665 JAD08627.1 AY842303 AAX44045.1 AB543754 BAI67740.1
Proteomes
UP000007151
UP000218220
UP000053268
UP000053240
UP000235965
UP000092461
+ More
UP000008237 UP000075809 UP000053825 UP000078540 UP000078541 UP000007755 UP000053105 UP000078492 UP000005205 UP000078542 UP000242457 UP000005203 UP000279307 UP000092462 UP000076502 UP000000311 UP000002320 UP000036403 UP000215335 UP000009046 UP000075882 UP000075902 UP000069272 UP000075920 UP000008820 UP000019118 UP000030742 UP000075885 UP000076407 UP000075886 UP000075903 UP000075883 UP000002358 UP000007266 UP000075840 UP000007062 UP000075884 UP000076408 UP000075900 UP000030765 UP000075881 UP000075880 UP000183832 UP000000673 UP000015103 UP000069940 UP000249989
UP000008237 UP000075809 UP000053825 UP000078540 UP000078541 UP000007755 UP000053105 UP000078492 UP000005205 UP000078542 UP000242457 UP000005203 UP000279307 UP000092462 UP000076502 UP000000311 UP000002320 UP000036403 UP000215335 UP000009046 UP000075882 UP000075902 UP000069272 UP000075920 UP000008820 UP000019118 UP000030742 UP000075885 UP000076407 UP000075886 UP000075903 UP000075883 UP000002358 UP000007266 UP000075840 UP000007062 UP000075884 UP000076408 UP000075900 UP000030765 UP000075881 UP000075880 UP000183832 UP000000673 UP000015103 UP000069940 UP000249989
Interpro
Gene 3D
ProteinModelPortal
A0A2H1VR20
A0A1E1WCW9
A0A2W1BNQ5
G0YQM3
A0A1V0EMB5
A0A212EGJ4
+ More
A0A0N7BAL6 Q3ZTR5 A0A1E1WUU5 A0A2A4IWA8 A0A194PGW1 A0A194QLE2 Q6VRU6 A0A2J7QFH5 E0D6T2 L8B0S6 A0A1B0GJJ1 E2C765 A0A151WTM8 A0A336KII5 A0A068Q7V1 A0A310SLY2 A0A0L7RH10 A0A195BTA5 A0A0H4BGL7 A0A195FJ10 F4WUR0 L7Y4E9 A0A0M9A7L2 A0A195DM36 A0A158P2D5 A0A195D379 A0A2A3EIL5 A0A088A9H3 A0A3L8E090 A0A1B0D5T7 A0A1L8DH51 A0A1L8DGZ4 A0A154PGQ5 E2AY34 B0W3Y0 A0A0J7L871 A0A1L5YQJ2 A0A232EFE1 E0VT39 A0A182L2Y8 A0A182U1Q0 A0A182FCB7 H7CI07 U5EHU6 A0A182WKT4 A0A1B6CYF0 A0A1S4FWV2 Q16LQ2 A9XCF0 A0A2M3ZFQ8 N6UMH2 U4U4S1 A0A182PMI9 A0A182XMS6 A0A182Q7X5 A0A182VHW3 A0A182LWI6 A0A0F7DCF2 K7JC66 D6WIQ8 A0A182HWN3 Q7Q6R8 A0A182NIR3 A0A182YD49 A0A1Y1M421 A0A182RZS8 A0A084WA50 A0A182KDN3 A0A182IKV4 R4WQ03 A0A1J1I6S5 A0A2M4BB90 A0A2M4A2Y8 A0A2M4BB67 W5JIJ2 A0A1Q3G4C9 A0A1Q3G4C5 A0A2M3YYP5 A0A2M4A4J9 T1HDJ0 A0A1Q3G490 A0A1Q3G4K0 A0A023FAB5 A0A182H8U9 A0A146LMC7 A0A0K8SX26 A0A0A9W299 A0A1B0YYT9 E9IE69 A0A2S0X9U7 A0A0A1XCF3 Q2KPA5 D2Z1J8
A0A0N7BAL6 Q3ZTR5 A0A1E1WUU5 A0A2A4IWA8 A0A194PGW1 A0A194QLE2 Q6VRU6 A0A2J7QFH5 E0D6T2 L8B0S6 A0A1B0GJJ1 E2C765 A0A151WTM8 A0A336KII5 A0A068Q7V1 A0A310SLY2 A0A0L7RH10 A0A195BTA5 A0A0H4BGL7 A0A195FJ10 F4WUR0 L7Y4E9 A0A0M9A7L2 A0A195DM36 A0A158P2D5 A0A195D379 A0A2A3EIL5 A0A088A9H3 A0A3L8E090 A0A1B0D5T7 A0A1L8DH51 A0A1L8DGZ4 A0A154PGQ5 E2AY34 B0W3Y0 A0A0J7L871 A0A1L5YQJ2 A0A232EFE1 E0VT39 A0A182L2Y8 A0A182U1Q0 A0A182FCB7 H7CI07 U5EHU6 A0A182WKT4 A0A1B6CYF0 A0A1S4FWV2 Q16LQ2 A9XCF0 A0A2M3ZFQ8 N6UMH2 U4U4S1 A0A182PMI9 A0A182XMS6 A0A182Q7X5 A0A182VHW3 A0A182LWI6 A0A0F7DCF2 K7JC66 D6WIQ8 A0A182HWN3 Q7Q6R8 A0A182NIR3 A0A182YD49 A0A1Y1M421 A0A182RZS8 A0A084WA50 A0A182KDN3 A0A182IKV4 R4WQ03 A0A1J1I6S5 A0A2M4BB90 A0A2M4A2Y8 A0A2M4BB67 W5JIJ2 A0A1Q3G4C9 A0A1Q3G4C5 A0A2M3YYP5 A0A2M4A4J9 T1HDJ0 A0A1Q3G490 A0A1Q3G4K0 A0A023FAB5 A0A182H8U9 A0A146LMC7 A0A0K8SX26 A0A0A9W299 A0A1B0YYT9 E9IE69 A0A2S0X9U7 A0A0A1XCF3 Q2KPA5 D2Z1J8
PDB
4F3L
E-value=1.44415e-35,
Score=369
Ontologies
GO
Topology
Subcellular location
Nucleus
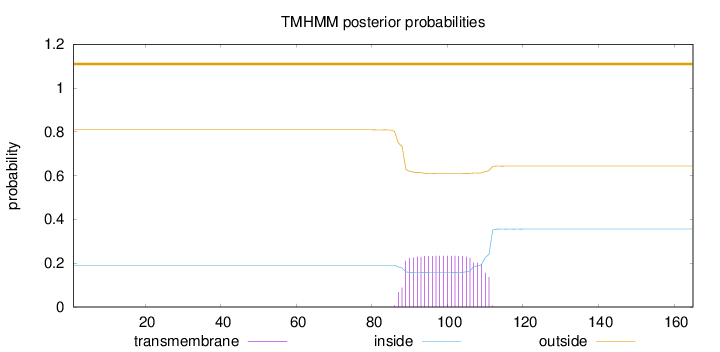
Length:
165
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
5.20316
Exp number, first 60 AAs:
0.00036
Total prob of N-in:
0.19086
outside
1 - 165
Population Genetic Test Statistics
Pi
1.631529
Theta
7.537294
Tajima's D
-2.479653
CLR
0.180736
CSRT
0.000349982500874956
Interpretation
Uncertain
