Gene
KWMTBOMO00390 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000504
Annotation
Ankyrin_repeat_domain-containing_protein_11_[Papilio_machaon]
Location in the cell
PlasmaMembrane Reliability : 4.122
Sequence
CDS
ATGGAGACTACTAGAGCGGAGATGGAAACGTGGTTGGATTGGTGCGTCCGAAAAAATGACATCAAGCAATTGTGGGCTGCCCAACCTACATATATAATATCTCAGGCCGTTTACGTTTTGGCTGGCCTCTTAACATTATTTCATGCTTTTAAAAAAGGCGGAAGATGGCCTTATTTTTGGCTCGGTACCGTACTTCATGGGATTTACGCAGATAACTTCTGGCATTTCGTATTGCCAGAGTACGATAATTTCTGGCATTCGCAGACCCCTATCATATTCCTGGGAGCCCGGCTACCCCTACATATAATTTTGTTGTATCCCGCATTTATTTACCACGCAGCGTATGCAGTTTCAAAGCTGAATTTTCCACGTTACGCGGAGCCTTTCGCGGTGGGCTTGGTTACTGTACTGATAGATATTCCCTACGACATTGTGGCGGTCAAGTTCGTACACTGGACCTGGCACGACACTGACCCTAACATATTCGATCGTCACTACTGGGTTCCCTGGAACTCCTATTACTTCCACGCTACATTCACAGCAAGCTTCTTCTACTTCTTTACTGCGATCAGAAATTGGTATGCGCCGAAAGTTGGACAGTGGGCTTCTGCGGGCAAAAAGGTGGAATGGAAAACGTTAATTCTAGCAACGATCCTGGGGATGCCAGGCGGTGTACTCCTCTTCGTACCGATCTATCACCCGCTCCACGACATATACAAAATACATTCAGAGATCACATTCTTCCTTTTGTTTGCGATTTATGGCACTTTCGTGATCCACGGGCTGCTTAGCGATAGAGAGAAATCCAAGGAAAAACTTACTCTTATAGATTACATCCTGATACTGCAACTGGCCCTCCACTACGTAATATATTGGATATTTGTTGTGTTCTTCCACCCAGAGAGGGAATGGTCTACCGGACTGCACGAGCCGGTCGGTCCATGCAACGAAGTAGCTAGTTTAGTCACGCCTTTTGGACAGACCTTGTACAAACGTAAGTACTTCTGCCCGGACCAGTACGACGAGGGTTACTTTGACTTCCACTGCGTCGGCGGCAAGAAGCCTCCAAACGGTGCTACTTGGTACACTATTTGTGGTACGCCATTCACGAATCGTGCTGAATATATTGTGCTGCTAACAACGATCCTCTTCCTAGCCGCCGGCGTCTTCTACGGCCTTTACTTCAGAACTATTCAGTCTGTACCAGTACCAACTAAGAAGCAGAAAACCAAATAA
Protein
METTRAEMETWLDWCVRKNDIKQLWAAQPTYIISQAVYVLAGLLTLFHAFKKGGRWPYFWLGTVLHGIYADNFWHFVLPEYDNFWHSQTPIIFLGARLPLHIILLYPAFIYHAAYAVSKLNFPRYAEPFAVGLVTVLIDIPYDIVAVKFVHWTWHDTDPNIFDRHYWVPWNSYYFHATFTASFFYFFTAIRNWYAPKVGQWASAGKKVEWKTLILATILGMPGGVLLFVPIYHPLHDIYKIHSEITFFLLFAIYGTFVIHGLLSDREKSKEKLTLIDYILILQLALHYVIYWIFVVFFHPEREWSTGLHEPVGPCNEVASLVTPFGQTLYKRKYFCPDQYDEGYFDFHCVGGKKPPNGATWYTICGTPFTNRAEYIVLLTTILFLAAGVFYGLYFRTIQSVPVPTKKQKTK
Summary
Uniprot
A0A3G1T1B0
A0A1E1WTQ3
A0A194QLW0
A0A194PF94
H9ITC4
S4PFG5
+ More
A0A2H1WY50 A0A212EGI7 A0A0L7LF11 A0A1B1MRN2 A0A2J7QRR2 A0A1B6FRE8 D6WER5 A0A067RDJ7 A0A164MD87 A0A0P5QV78 A0A0P6FLV5 A0A0P4XQ16 A0A0P5CLN8 A0A0P6CLK5 A0A1B6CE90 A0A0P5JUZ1 J3JXI2 N6TP16 U4UEX1 A0A2J7QRS7 A0A0P5BKY2 A0A0P5Z662 A0A1S3DUD5 A0A1S4FSW1 A0A0P4X0G4 A0A226DL46 Q16QQ2 A0A0P4YTC6 A0A0P5EBX2 A0A182GWC5 A0A1D2MVH5 A0A0N8EN25 A0A1B0EZ80 A0A1S3HH07 A0A0P5XT26 A0A1W4XFU8 A0A0P4WS11 A0A1S3HFG2 A0A0B7AIH8 V4A2B2 B0WZP5 A0A3Q0JLW7 A0A210PLZ9 A0A0P5BSA5 A7RXC1 A0A1B0DA53 A0A3Q0JPZ7 A0A1B6I261 A0A1I7SM79 A0A1W2WEU8 A0A0P5CEH0 A0A0N8BES4 A0A2T7PWT0 H2Z7V6 A0A0P5DIG0 F6S414 A0A260ZF91 E3MAX2 A0A261CMC7 H3DSH7 G0MGX0 A0A1I7TZT5 A0A2H2HW93 A0A2G5SL41 A8XP16 A0A2C9JE79 Q23660 Q16QQ0 A0A087THA2 C3ZJ16 C3YH82 A0A1L8D733 A0A182N7V2 A0A182PB68 A0A182VL76 A0A1I8ARY9 A0A016WR98 A0A182K8R6 A0A016WS70 A0A183G1K3 A0A3P7ZI83 R7TS47 A0A084WEX0 A0A368GMA5 A0A182WAM2 A0A0D6LP32 A0A182IUM2 A0A2S2NYS8 A0NEB3 A0A2A2LHS0
A0A2H1WY50 A0A212EGI7 A0A0L7LF11 A0A1B1MRN2 A0A2J7QRR2 A0A1B6FRE8 D6WER5 A0A067RDJ7 A0A164MD87 A0A0P5QV78 A0A0P6FLV5 A0A0P4XQ16 A0A0P5CLN8 A0A0P6CLK5 A0A1B6CE90 A0A0P5JUZ1 J3JXI2 N6TP16 U4UEX1 A0A2J7QRS7 A0A0P5BKY2 A0A0P5Z662 A0A1S3DUD5 A0A1S4FSW1 A0A0P4X0G4 A0A226DL46 Q16QQ2 A0A0P4YTC6 A0A0P5EBX2 A0A182GWC5 A0A1D2MVH5 A0A0N8EN25 A0A1B0EZ80 A0A1S3HH07 A0A0P5XT26 A0A1W4XFU8 A0A0P4WS11 A0A1S3HFG2 A0A0B7AIH8 V4A2B2 B0WZP5 A0A3Q0JLW7 A0A210PLZ9 A0A0P5BSA5 A7RXC1 A0A1B0DA53 A0A3Q0JPZ7 A0A1B6I261 A0A1I7SM79 A0A1W2WEU8 A0A0P5CEH0 A0A0N8BES4 A0A2T7PWT0 H2Z7V6 A0A0P5DIG0 F6S414 A0A260ZF91 E3MAX2 A0A261CMC7 H3DSH7 G0MGX0 A0A1I7TZT5 A0A2H2HW93 A0A2G5SL41 A8XP16 A0A2C9JE79 Q23660 Q16QQ0 A0A087THA2 C3ZJ16 C3YH82 A0A1L8D733 A0A182N7V2 A0A182PB68 A0A182VL76 A0A1I8ARY9 A0A016WR98 A0A182K8R6 A0A016WS70 A0A183G1K3 A0A3P7ZI83 R7TS47 A0A084WEX0 A0A368GMA5 A0A182WAM2 A0A0D6LP32 A0A182IUM2 A0A2S2NYS8 A0NEB3 A0A2A2LHS0
Pubmed
EMBL
MG846913
AXY94765.1
GDQN01010419
GDQN01000702
JAT80635.1
JAT90352.1
+ More
KQ461196 KPJ06349.1 KQ459605 KPI91952.1 BABH01007199 BABH01007200 GAIX01004017 JAA88543.1 ODYU01011944 SOQ57978.1 AGBW02015069 OWR40598.1 JTDY01001401 KOB73969.1 KU529968 ANS71233.1 NEVH01011885 PNF31282.1 GECZ01016984 JAS52785.1 KQ971326 EEZ99870.2 KK852529 KDR21951.1 LRGB01003024 KZS04952.1 GDIQ01109240 JAL42486.1 GDIQ01046278 JAN48459.1 GDIP01242308 JAI81093.1 GDIP01172438 JAJ50964.1 GDIQ01090647 JAN04090.1 GEDC01025532 JAS11766.1 GDIQ01193308 JAK58417.1 BT127951 AEE62913.1 APGK01027030 APGK01027031 APGK01027032 KB740648 ENN79763.1 KB632292 ERL91587.1 PNF31280.1 GDIP01183131 JAJ40271.1 GDIP01048609 JAM55106.1 GDIP01252219 JAI71182.1 LNIX01000017 OXA45718.1 CH477738 EAT36725.1 GDIP01222837 JAJ00565.1 GDIP01145122 JAJ78280.1 JXUM01004303 KQ560172 KXJ84008.1 LJIJ01000501 ODM96804.1 GDIQ01010668 JAN84069.1 AJWK01021990 GDIP01067699 JAM36016.1 GDIP01252424 JAI70977.1 HACG01033743 CEK80608.1 KB202917 ESO87411.1 DS232212 EDS37720.1 NEDP02005588 OWF37505.1 GDIP01181488 JAJ41914.1 DS469549 EDO43939.1 AJVK01028830 GECU01026761 JAS80945.1 GDIP01172437 JAJ50965.1 GDIQ01184867 JAK66858.1 PZQS01000001 PVD37869.1 GDIP01172439 JAJ50963.1 EAAA01001891 NMWX01000163 OZF84242.1 DS268432 LFJK02000005 EFO97428.1 POM49539.1 NIPN01000003 OZG23195.1 GL379794 EGT57999.1 PDUG01000006 PIC15737.1 HE600961 CAP34256.1 BX284606 CAA85502.1 EAT36727.1 KK115224 KFM64491.1 GG666631 EEN47445.1 GG666513 EEN60318.1 GFDF01011833 JAV02251.1 JARK01000142 EYC42106.1 EYC42107.1 UZAH01028684 VDP01717.1 AMQN01011295 KB308811 ELT96479.1 ATLV01023260 KE525341 KFB48764.1 JOJR01000100 RCN45501.1 KE124970 EPB73810.1 GGMR01009706 MBY22325.1 AAAB01008960 EAU76432.2 LIAE01006728 PAV85806.1
KQ461196 KPJ06349.1 KQ459605 KPI91952.1 BABH01007199 BABH01007200 GAIX01004017 JAA88543.1 ODYU01011944 SOQ57978.1 AGBW02015069 OWR40598.1 JTDY01001401 KOB73969.1 KU529968 ANS71233.1 NEVH01011885 PNF31282.1 GECZ01016984 JAS52785.1 KQ971326 EEZ99870.2 KK852529 KDR21951.1 LRGB01003024 KZS04952.1 GDIQ01109240 JAL42486.1 GDIQ01046278 JAN48459.1 GDIP01242308 JAI81093.1 GDIP01172438 JAJ50964.1 GDIQ01090647 JAN04090.1 GEDC01025532 JAS11766.1 GDIQ01193308 JAK58417.1 BT127951 AEE62913.1 APGK01027030 APGK01027031 APGK01027032 KB740648 ENN79763.1 KB632292 ERL91587.1 PNF31280.1 GDIP01183131 JAJ40271.1 GDIP01048609 JAM55106.1 GDIP01252219 JAI71182.1 LNIX01000017 OXA45718.1 CH477738 EAT36725.1 GDIP01222837 JAJ00565.1 GDIP01145122 JAJ78280.1 JXUM01004303 KQ560172 KXJ84008.1 LJIJ01000501 ODM96804.1 GDIQ01010668 JAN84069.1 AJWK01021990 GDIP01067699 JAM36016.1 GDIP01252424 JAI70977.1 HACG01033743 CEK80608.1 KB202917 ESO87411.1 DS232212 EDS37720.1 NEDP02005588 OWF37505.1 GDIP01181488 JAJ41914.1 DS469549 EDO43939.1 AJVK01028830 GECU01026761 JAS80945.1 GDIP01172437 JAJ50965.1 GDIQ01184867 JAK66858.1 PZQS01000001 PVD37869.1 GDIP01172439 JAJ50963.1 EAAA01001891 NMWX01000163 OZF84242.1 DS268432 LFJK02000005 EFO97428.1 POM49539.1 NIPN01000003 OZG23195.1 GL379794 EGT57999.1 PDUG01000006 PIC15737.1 HE600961 CAP34256.1 BX284606 CAA85502.1 EAT36727.1 KK115224 KFM64491.1 GG666631 EEN47445.1 GG666513 EEN60318.1 GFDF01011833 JAV02251.1 JARK01000142 EYC42106.1 EYC42107.1 UZAH01028684 VDP01717.1 AMQN01011295 KB308811 ELT96479.1 ATLV01023260 KE525341 KFB48764.1 JOJR01000100 RCN45501.1 KE124970 EPB73810.1 GGMR01009706 MBY22325.1 AAAB01008960 EAU76432.2 LIAE01006728 PAV85806.1
Proteomes
UP000053240
UP000053268
UP000005204
UP000007151
UP000037510
UP000235965
+ More
UP000007266 UP000027135 UP000076858 UP000019118 UP000030742 UP000079169 UP000198287 UP000008820 UP000069940 UP000249989 UP000094527 UP000092461 UP000085678 UP000192223 UP000030746 UP000002320 UP000242188 UP000001593 UP000092462 UP000095284 UP000245119 UP000007875 UP000008144 UP000216624 UP000008281 UP000237256 UP000216463 UP000005239 UP000008068 UP000095282 UP000005237 UP000230233 UP000008549 UP000076420 UP000001940 UP000054359 UP000001554 UP000075884 UP000075885 UP000075903 UP000095287 UP000024635 UP000075881 UP000050761 UP000014760 UP000030765 UP000252519 UP000075920 UP000075880 UP000007062 UP000218231
UP000007266 UP000027135 UP000076858 UP000019118 UP000030742 UP000079169 UP000198287 UP000008820 UP000069940 UP000249989 UP000094527 UP000092461 UP000085678 UP000192223 UP000030746 UP000002320 UP000242188 UP000001593 UP000092462 UP000095284 UP000245119 UP000007875 UP000008144 UP000216624 UP000008281 UP000237256 UP000216463 UP000005239 UP000008068 UP000095282 UP000005237 UP000230233 UP000008549 UP000076420 UP000001940 UP000054359 UP000001554 UP000075884 UP000075885 UP000075903 UP000095287 UP000024635 UP000075881 UP000050761 UP000014760 UP000030765 UP000252519 UP000075920 UP000075880 UP000007062 UP000218231
PRIDE
Pfam
Interpro
IPR001841
Znf_RING
+ More
IPR000967 Znf_NFX1
IPR019786 Zinc_finger_PHD-type_CS
IPR034078 NFX1_fam
IPR036249 Thioredoxin-like_sf
IPR002109 Glutaredoxin
IPR013766 Thioredoxin_domain
IPR033658 GRX_PICOT-like
IPR001810 F-box_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR006603 PQ-loop_rpt
IPR036047 F-box-like_dom_sf
IPR020472 G-protein_beta_WD-40_rep
IPR036322 WD40_repeat_dom_sf
IPR019775 WD40_repeat_CS
IPR000967 Znf_NFX1
IPR019786 Zinc_finger_PHD-type_CS
IPR034078 NFX1_fam
IPR036249 Thioredoxin-like_sf
IPR002109 Glutaredoxin
IPR013766 Thioredoxin_domain
IPR033658 GRX_PICOT-like
IPR001810 F-box_dom
IPR015943 WD40/YVTN_repeat-like_dom_sf
IPR001680 WD40_repeat
IPR017986 WD40_repeat_dom
IPR006603 PQ-loop_rpt
IPR036047 F-box-like_dom_sf
IPR020472 G-protein_beta_WD-40_rep
IPR036322 WD40_repeat_dom_sf
IPR019775 WD40_repeat_CS
Gene 3D
ProteinModelPortal
A0A3G1T1B0
A0A1E1WTQ3
A0A194QLW0
A0A194PF94
H9ITC4
S4PFG5
+ More
A0A2H1WY50 A0A212EGI7 A0A0L7LF11 A0A1B1MRN2 A0A2J7QRR2 A0A1B6FRE8 D6WER5 A0A067RDJ7 A0A164MD87 A0A0P5QV78 A0A0P6FLV5 A0A0P4XQ16 A0A0P5CLN8 A0A0P6CLK5 A0A1B6CE90 A0A0P5JUZ1 J3JXI2 N6TP16 U4UEX1 A0A2J7QRS7 A0A0P5BKY2 A0A0P5Z662 A0A1S3DUD5 A0A1S4FSW1 A0A0P4X0G4 A0A226DL46 Q16QQ2 A0A0P4YTC6 A0A0P5EBX2 A0A182GWC5 A0A1D2MVH5 A0A0N8EN25 A0A1B0EZ80 A0A1S3HH07 A0A0P5XT26 A0A1W4XFU8 A0A0P4WS11 A0A1S3HFG2 A0A0B7AIH8 V4A2B2 B0WZP5 A0A3Q0JLW7 A0A210PLZ9 A0A0P5BSA5 A7RXC1 A0A1B0DA53 A0A3Q0JPZ7 A0A1B6I261 A0A1I7SM79 A0A1W2WEU8 A0A0P5CEH0 A0A0N8BES4 A0A2T7PWT0 H2Z7V6 A0A0P5DIG0 F6S414 A0A260ZF91 E3MAX2 A0A261CMC7 H3DSH7 G0MGX0 A0A1I7TZT5 A0A2H2HW93 A0A2G5SL41 A8XP16 A0A2C9JE79 Q23660 Q16QQ0 A0A087THA2 C3ZJ16 C3YH82 A0A1L8D733 A0A182N7V2 A0A182PB68 A0A182VL76 A0A1I8ARY9 A0A016WR98 A0A182K8R6 A0A016WS70 A0A183G1K3 A0A3P7ZI83 R7TS47 A0A084WEX0 A0A368GMA5 A0A182WAM2 A0A0D6LP32 A0A182IUM2 A0A2S2NYS8 A0NEB3 A0A2A2LHS0
A0A2H1WY50 A0A212EGI7 A0A0L7LF11 A0A1B1MRN2 A0A2J7QRR2 A0A1B6FRE8 D6WER5 A0A067RDJ7 A0A164MD87 A0A0P5QV78 A0A0P6FLV5 A0A0P4XQ16 A0A0P5CLN8 A0A0P6CLK5 A0A1B6CE90 A0A0P5JUZ1 J3JXI2 N6TP16 U4UEX1 A0A2J7QRS7 A0A0P5BKY2 A0A0P5Z662 A0A1S3DUD5 A0A1S4FSW1 A0A0P4X0G4 A0A226DL46 Q16QQ2 A0A0P4YTC6 A0A0P5EBX2 A0A182GWC5 A0A1D2MVH5 A0A0N8EN25 A0A1B0EZ80 A0A1S3HH07 A0A0P5XT26 A0A1W4XFU8 A0A0P4WS11 A0A1S3HFG2 A0A0B7AIH8 V4A2B2 B0WZP5 A0A3Q0JLW7 A0A210PLZ9 A0A0P5BSA5 A7RXC1 A0A1B0DA53 A0A3Q0JPZ7 A0A1B6I261 A0A1I7SM79 A0A1W2WEU8 A0A0P5CEH0 A0A0N8BES4 A0A2T7PWT0 H2Z7V6 A0A0P5DIG0 F6S414 A0A260ZF91 E3MAX2 A0A261CMC7 H3DSH7 G0MGX0 A0A1I7TZT5 A0A2H2HW93 A0A2G5SL41 A8XP16 A0A2C9JE79 Q23660 Q16QQ0 A0A087THA2 C3ZJ16 C3YH82 A0A1L8D733 A0A182N7V2 A0A182PB68 A0A182VL76 A0A1I8ARY9 A0A016WR98 A0A182K8R6 A0A016WS70 A0A183G1K3 A0A3P7ZI83 R7TS47 A0A084WEX0 A0A368GMA5 A0A182WAM2 A0A0D6LP32 A0A182IUM2 A0A2S2NYS8 A0NEB3 A0A2A2LHS0
Ontologies
GO
PANTHER
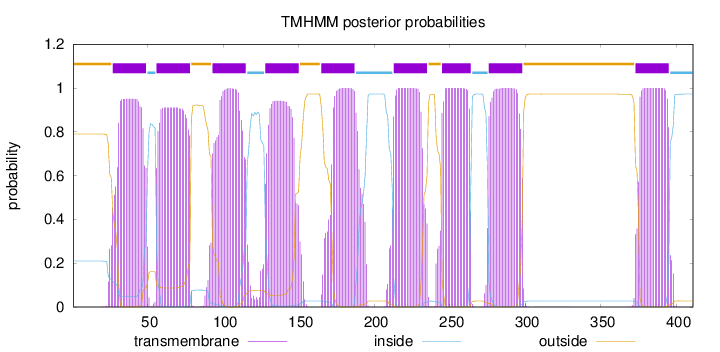
Topology
Length:
411
Number of predicted TMHs:
9
Exp number of AAs in TMHs:
191.46409
Exp number, first 60 AAs:
24.36879
Total prob of N-in:
0.21016
POSSIBLE N-term signal
sequence
outside
1 - 26
TMhelix
27 - 49
inside
50 - 55
TMhelix
56 - 78
outside
79 - 92
TMhelix
93 - 115
inside
116 - 127
TMhelix
128 - 150
outside
151 - 164
TMhelix
165 - 187
inside
188 - 212
TMhelix
213 - 235
outside
236 - 244
TMhelix
245 - 264
inside
265 - 275
TMhelix
276 - 298
outside
299 - 372
TMhelix
373 - 395
inside
396 - 411
Population Genetic Test Statistics
Pi
31.133177
Theta
31.360644
Tajima's D
0.185715
CLR
0.07419
CSRT
0.426478676066197
Interpretation
Uncertain
