Gene
KWMTBOMO00388
Pre Gene Modal
BGIBMGA000719
Annotation
PREDICTED:_membrane-bound_transcription_factor_site-1_protease_[Amyelois_transitella]
Full name
Membrane-bound transcription factor site-1 protease
Alternative Name
Endopeptidase S1P
Subtilisin/kexin-isozyme 1
Subtilisin/kexin-isozyme 1
Location in the cell
PlasmaMembrane Reliability : 1.048
Sequence
CDS
ATGGGGTTCACCCATACAATTCTTCTAAGTGTAATGCTTTATCTCCCTATCTTATTAATTTCTTCTAAAGAAGTGCGTGAAAAAAATGAAAATTGCAATACCACTGAGTCTGAACGTGTTCAATACGAGTTCACTACCGATGTTGTTAAAAGTCAGCACATAATAACATTTAAAGGATACTATCCCAGGAGCACAAGAGAAAATTATGTAAATGCCGCTCTAACCAGTGCAGGTATAACAGACTGGAAGATTATTAAACGCAACAATCCAGCCAAAGAATATCCTAGTGACTTTGATGTTGTTGTCCTAGAAAATGCAGGAAGAAAAGCATTGGAAGCCCTGCAAGATCACCCAGCAGTTCGACGGGTCACAGCCCAACGTCAAGTCACACGGTCCATAAAGTATGTTAAAGAGGATGAGTGTGGATCAGGGGGCTGTTTGTATTCTGGGTGGCGGAATTATCGCGGAAGAGGACTGCACTCTCTTCGAAAAACTCGTGACAATGGATACACGTCACGCAAACTATTACGGACTGTCCCGCGCCAGATCACCTCTGTGTTGAAAGCAGACGTTCTTTGGTCTCTTGGTGTCACAGGTGAAGGTATAAAGGTCGCCGTGTTTGATACCGGCTTAGCTCGTAACCATCCTCACTTTGGGCGGATCCGTGAACGTACCGACTGGACGGGAGAAGACACATTGGACGACGCCTTAGGCCACGGAACGTTCGTTGCCGGAGTAATAGCTTCGAAGGCCGACTGTTTGGGATTCGCTCCCGACGCCGATTTGCACATATTCCGAGTATTCACTGATAATCAGGTTTCATACACTTCGTGGTTCCTGGACGCGTTCAACTACGCAATAATGCGCAAAATAGACGTTCTCAATCTCAGCATCGGGGGACCGGATTTTATGGATCATCCGTTTGTCGACAAAGTCTGGGAACTCAGTGCTAATAAGGTAATTATGGTATCGGCGATCGGTAACGACGGTCCTCTGTACGGCACACTGAACAACCCCGCTGATCAGATGGACGTCATAGGTGTCGGCGGTATAGGGTTCGATGACCGCATCGCGAAATTTTCGTCGAGAGGAATGACAACATGGGAGTTACCACAAGGTTACGGTCGTATGAAGCCCGACATTGTAACGTACGGCAGTGGAGTCCGTGGATCGAGCGTGAATGGTGGATGTAAATCACTTAGCGGTACATCGGTAGCGTCGCCGGTTGTAGCTGGCGCCATAGCGTTACTAGCGAGCGGTGTTCCACGTCAGAATTTAACTCCGGCATCTGTTAAACAAGCCTTATGCAGCACAGCGCTAAGACTACAAGGGCCTAACATGTTCGAACAGGGACACGGGAAATTGGACCTTATCAGCGCTTATCAGTTCCTCAGGAAGTACGAGCCGCAAGCTACATTAAGCCCGAATTACATTGACCTCACTGAATGTCAGTACATGTGGCCGTACTGCACTCAGCCGCTGTATTACAGCTCGCAGCCAACGATCGCGAACGTGACCGTCATTAATGGACTGAGCGTCGTTGGGAATGTGAAAGAAGTCTCGTGGCATCCACATCTGCCCCATGGCGTTCTACTCTCGGTGTCTTCAGATTACAGCGATGTCCTCTGGCCTTGGTCTGGCTGGCTGGCCCTCAGCTTCACAGTCCGGGAAGAAGGGGCTAACTTTGATGGTATCATCGAGGGTCACGTGAACGTAACGATTGAAAGTCACGACGATACAGGTGATAGAACAATAAAGAACGCCACACTCACATTGCCGATACGAGCTCGAGTGATCCCAGTTCCGGTGCGATCCCGGCGTCTCCTCTGGGACCAGTTCCACAGTCTACGTTACCCCGGCGGCTACTTCCCTAGGGACGATCTACGAGCCAAACACGATCCCCTCGACTGGCACGCCGATCATATACACACCAATTTTAGGGACATGTACAGGAGATTGCGCGAACACGGATTCTACGTTGAAGTCATGGGTACACCCTTAACATGTATCGACACGTCGCTCTACGGCGCCCTGTTACTCGTTGACCCCGAAGATGAATATTTCCCCGAAGAGATGGCAACATTGAAGAAAGCCGTTGACGCTGGACTATCGTTAATTGTTTTCGCCGATTGGTACAACGCTTCCCTTCTGCGGCACGTGAAGTTTTACGACGAAAACACAAGGCAATGGTGGATTCCTGAGACGGGAGGATCAAACGTTCCCGCTCTGAACGATTTACTCAGCATGTATCAGGTGGCGCTAGGCGATAGAGTATTTGAAGGTTCATTCAAACTGGGCGGACACCCGATGTATTACGCGAGCGGAACTCACATACACAGCTTCCCTGAACACGGTGTACTGATCACGGCCCCTCTCAACGATCAAGGGCAACAGATAATATCAGGTGAAAAGCCGGGGGCCGCGAACGGCAAAACGGAAGTCACACAACCGTTGCAAGTCCCGATCCTAGGTCTCCTACAAACTGATGGAGATATTCATGACTATTCAAACGATACCGTGGATAAAGTACCTAAGTCTGGCCGTCTGGTCGTTTACGGAGATTCATCTTGTCTGGAAGGTGGAACTGCCCGGAACTGTCACTGGTTGCTTTTGGCCGCTTTACAATACGCAATAGTGGGTCACGTGCCTCCAGCTCTCAAAGAAGCCGCAACGCCGCAAGAGAAACGCGTCAGAGATGTGCATATAATCCCTTCAGAACTGCCTCGACGTGCCGAAGGGGGACGTCTCCATGTCTACTCTCGCGTCCTATCGCCCGATGGAAGTGGACCACGTCCAATACCAGAATGCTTCAGTCCCACCCCGCTACCGTCCCAGCCAGTACACGCTGCTCCATCTGCCAGGACATTGGCTCCTAGACATCAGCCTACAGATCCTAAGAGTATCGGTCCTCCAGAAATAGAAGGCACCGAACCAGCGCCCAGAGCGTGGCGAGGAGCAGGCGCCGCGCCTTCCAGAGTTCACCCGCAGTCGGAAACGACACCCACTAGTCTAACGAACCGAGCTATCACACTACTAGCGATCGTAACTGCTATATACACTGTCGCTCAGCTGTGGAAGCGGTGGGGCCGCAGGATCCGGAGACGCAGAATCATCTCTCTAGCCACCTAG
Protein
MGFTHTILLSVMLYLPILLISSKEVREKNENCNTTESERVQYEFTTDVVKSQHIITFKGYYPRSTRENYVNAALTSAGITDWKIIKRNNPAKEYPSDFDVVVLENAGRKALEALQDHPAVRRVTAQRQVTRSIKYVKEDECGSGGCLYSGWRNYRGRGLHSLRKTRDNGYTSRKLLRTVPRQITSVLKADVLWSLGVTGEGIKVAVFDTGLARNHPHFGRIRERTDWTGEDTLDDALGHGTFVAGVIASKADCLGFAPDADLHIFRVFTDNQVSYTSWFLDAFNYAIMRKIDVLNLSIGGPDFMDHPFVDKVWELSANKVIMVSAIGNDGPLYGTLNNPADQMDVIGVGGIGFDDRIAKFSSRGMTTWELPQGYGRMKPDIVTYGSGVRGSSVNGGCKSLSGTSVASPVVAGAIALLASGVPRQNLTPASVKQALCSTALRLQGPNMFEQGHGKLDLISAYQFLRKYEPQATLSPNYIDLTECQYMWPYCTQPLYYSSQPTIANVTVINGLSVVGNVKEVSWHPHLPHGVLLSVSSDYSDVLWPWSGWLALSFTVREEGANFDGIIEGHVNVTIESHDDTGDRTIKNATLTLPIRARVIPVPVRSRRLLWDQFHSLRYPGGYFPRDDLRAKHDPLDWHADHIHTNFRDMYRRLREHGFYVEVMGTPLTCIDTSLYGALLLVDPEDEYFPEEMATLKKAVDAGLSLIVFADWYNASLLRHVKFYDENTRQWWIPETGGSNVPALNDLLSMYQVALGDRVFEGSFKLGGHPMYYASGTHIHSFPEHGVLITAPLNDQGQQIISGEKPGAANGKTEVTQPLQVPILGLLQTDGDIHDYSNDTVDKVPKSGRLVVYGDSSCLEGGTARNCHWLLLAALQYAIVGHVPPALKEAATPQEKRVRDVHIIPSELPRRAEGGRLHVYSRVLSPDGSGPRPIPECFSPTPLPSQPVHAAPSARTLAPRHQPTDPKSIGPPEIEGTEPAPRAWRGAGAAPSRVHPQSETTPTSLTNRAITLLAIVTAIYTVAQLWKRWGRRIRRRRIISLAT
Summary
Description
Serine protease that catalyzes the first step in the proteolytic activation of the sterol regulatory element-binding proteins (SREBPs). Other known substrates are BDNF, GNPTAB and ATF6. Cleaves after hydrophobic or small residues, provided that Arg or Lys is in position P4. Cleaves known substrates after Arg-Ser-Val-Leu (SERBP-2), Arg-His-Leu-Leu (ATF6), Arg-Gly-Leu-Thr (BDNF) and its own propeptide after Arg-Arg-Leu-Leu. Mediates the protein cleavage of GNPTAB into subunit alpha and beta, thereby participating in biogenesis of lysosomes.
Catalytic Activity
Processes precursors containing basic and hydrophobic/aliphatic residues at P4 and P2, respectively, with a relatively relaxed acceptance of amino acids at P1 and P3.
Cofactor
Ca(2+)
Similarity
Belongs to the peptidase S8 family.
Keywords
Autocatalytic cleavage
Calcium
Cholesterol metabolism
Complete proteome
Direct protein sequencing
Endoplasmic reticulum
Glycoprotein
Golgi apparatus
Hydrolase
Lipid metabolism
Membrane
Phosphoprotein
Polymorphism
Protease
Reference proteome
Serine protease
Signal
Steroid metabolism
Sterol metabolism
Transmembrane
Transmembrane helix
Zymogen
Feature
chain Membrane-bound transcription factor site-1 protease
sequence variant In dbSNP:rs34701895.
sequence variant In dbSNP:rs34701895.
Uniprot
A0A2A4JC15
A0A212EGG8
A0A194PGX1
A0A194QLD6
A0A1Y1LMH9
A0A1Y1LPH7
+ More
D6WIB2 A0A139WLB7 A0A1W4WJX8 A0A1W4WIY9 A0A2J7PZP8 A0A2J7PZR5 A0A067QSV2 A0A0T6B9G1 A0A2A3EAP1 T1ISC2 A0A088A5Z3 A0A232EJF7 T1J4A8 A0A1L8GKG1 Q6AX98 A0A1L8GKI6 A0A1S3HYQ9 A0A154P3R4 Q0VA23 M3YC21 A0A2Y9K244 A0A384BZG3 A0A091S4F9 A0A0C9QMP8 F6S2F2 D2HRI4 A0A2Y9IG02 V9K9C3 A0A2Y9SY48 A0A384A5I7 G1KN46 A0A341AP56 Q08E55 A0A2Y9P8W9 A0A3L8SFL4 A0A2Y9SPV3 A0A2Y9NXL1 A0A099Z1P2 A0A093PUW3 A0A2Y9DNI6 A0A340X0E6 A0A2Y9K2N3 Q14703 F1PER6 A0A0P6JL18 S7MMG9 R0L9K5 A0A099ZYJ0 M3WFB1 G9K9U7 A0A151MLJ3 I3MBB7 G1RJD6 A0A286ZZH4 A0A2K5KZ34 A0A2U4BRP0 A0A1U7RFK3 A0A1U7R8F2 H0V6Z3 A0A2K6CMT2 A0A2K5V1G2 A0A2K5V1H8 A0A0D9S1D8 A0A2U4BRQ6 A0A096NPH3 H9ZES7 A0A2K5Z4Q0 H9G192 F7G9K2 A0A131YEJ8 A0A091GGY9 H0Z8S1 H2QBL9 E0VQL9 A0A2I0UCW7 A0A091N8D2 Q1LWH3 A0A1I9XGB5 A0A2K6K2W1 A0A2K6R346 A0A2K5IJ30 A0A131YQA9 A0A091VER6 A0A094KTH6 A0A1I9XGB3 G3SXF6 A0A091WI36 A0A1V4K102 A0A091N6X0 A0A091TEW2 L8I827 Q5R4I7 A0A2K5F5D8
D6WIB2 A0A139WLB7 A0A1W4WJX8 A0A1W4WIY9 A0A2J7PZP8 A0A2J7PZR5 A0A067QSV2 A0A0T6B9G1 A0A2A3EAP1 T1ISC2 A0A088A5Z3 A0A232EJF7 T1J4A8 A0A1L8GKG1 Q6AX98 A0A1L8GKI6 A0A1S3HYQ9 A0A154P3R4 Q0VA23 M3YC21 A0A2Y9K244 A0A384BZG3 A0A091S4F9 A0A0C9QMP8 F6S2F2 D2HRI4 A0A2Y9IG02 V9K9C3 A0A2Y9SY48 A0A384A5I7 G1KN46 A0A341AP56 Q08E55 A0A2Y9P8W9 A0A3L8SFL4 A0A2Y9SPV3 A0A2Y9NXL1 A0A099Z1P2 A0A093PUW3 A0A2Y9DNI6 A0A340X0E6 A0A2Y9K2N3 Q14703 F1PER6 A0A0P6JL18 S7MMG9 R0L9K5 A0A099ZYJ0 M3WFB1 G9K9U7 A0A151MLJ3 I3MBB7 G1RJD6 A0A286ZZH4 A0A2K5KZ34 A0A2U4BRP0 A0A1U7RFK3 A0A1U7R8F2 H0V6Z3 A0A2K6CMT2 A0A2K5V1G2 A0A2K5V1H8 A0A0D9S1D8 A0A2U4BRQ6 A0A096NPH3 H9ZES7 A0A2K5Z4Q0 H9G192 F7G9K2 A0A131YEJ8 A0A091GGY9 H0Z8S1 H2QBL9 E0VQL9 A0A2I0UCW7 A0A091N8D2 Q1LWH3 A0A1I9XGB5 A0A2K6K2W1 A0A2K6R346 A0A2K5IJ30 A0A131YQA9 A0A091VER6 A0A094KTH6 A0A1I9XGB3 G3SXF6 A0A091WI36 A0A1V4K102 A0A091N6X0 A0A091TEW2 L8I827 Q5R4I7 A0A2K5F5D8
EC Number
3.4.21.112
Pubmed
22118469
26354079
28004739
18362917
19820115
24845553
+ More
28648823 27762356 24813606 20431018 20010809 24402279 19393038 30282656 10944850 7788527 14702039 15616553 15489334 17974005 10644685 9990022 21719679 26091039 16341006 17975172 23236062 22293439 30723633 21993624 25319552 17431167 22002653 26830274 20360741 16136131 20566863 23594743 25362486 22751099
28648823 27762356 24813606 20431018 20010809 24402279 19393038 30282656 10944850 7788527 14702039 15616553 15489334 17974005 10644685 9990022 21719679 26091039 16341006 17975172 23236062 22293439 30723633 21993624 25319552 17431167 22002653 26830274 20360741 16136131 20566863 23594743 25362486 22751099
EMBL
NWSH01001943
PCG69615.1
AGBW02015069
OWR40595.1
KQ459605
KPI91954.1
+ More
KQ461196 KPJ06347.1 GEZM01051535 JAV74839.1 GEZM01051533 JAV74841.1 KQ971321 EFA00011.2 KYB28740.1 NEVH01020333 PNF21813.1 PNF21812.1 KK852981 KDR12950.1 LJIG01002956 KRT83984.1 KZ288305 PBC28785.1 JH431429 NNAY01004010 OXU18499.1 JH431841 CM004472 OCT84338.1 BC079695 AAH79695.1 OCT84339.1 KQ434809 KZC06556.1 BC121290 AAI21291.1 AEYP01035253 AEYP01035254 KK940306 KFQ50194.1 GBYB01001917 JAG71684.1 AAMC01102941 AAMC01102942 AAMC01102943 AAMC01102944 AAMC01102945 AAMC01102946 AAMC01102947 GL193222 EFB21511.1 JW861845 AFO94362.1 BC123412 AAI23413.1 QUSF01000023 RLW01270.1 KL887256 KGL74715.1 KL669222 KFW75862.1 D42053 AK291773 AC040169 BC114555 BC114961 AL133583 AAEX03004026 AAEX03004027 GEBF01001246 JAO02387.1 KE161800 EPQ05449.1 KB743829 EOA96927.1 KL870139 KGL86881.1 AANG04002512 JP013074 AES01672.1 AKHW03005795 KYO25431.1 AGTP01083736 AGTP01083737 AGTP01083738 ADFV01071441 ADFV01071442 ADFV01071443 AEMK02000039 DQIR01157021 DQIR01213789 DQIR01298853 HDB12498.1 AAKN02054945 AQIA01043817 AQIA01043818 AQIA01043819 AQIA01043820 AQIB01129880 AHZZ02019146 AHZZ02019147 JU477639 AFH34443.1 JU336898 JV048573 AFE80651.1 AFI38644.1 JSUE03026761 JSUE03026762 CM001272 EHH31903.1 GEDV01011589 JAP76968.1 KL447421 KFO73387.1 ABQF01035918 AACZ04051371 AC195663 GABC01003307 GABF01005018 GABD01002378 GABE01010437 NBAG03000295 JAA08031.1 JAA17127.1 JAA30722.1 JAA34302.1 PNI45721.1 DS235430 EEB15675.1 KZ505864 PKU43898.1 KL378584 KFP85686.1 BX548157 CT025908 CAN88262.1 KX013132 KX013134 KX013138 KX013139 KX013140 KX013142 KX013143 KX013144 APA62631.1 APA62633.1 APA62637.1 APA62638.1 APA62639.1 APA62641.1 APA62642.1 APA62643.1 GEDV01008281 JAP80276.1 KL410965 KFR01634.1 KL343431 KFZ54330.1 KX013135 KX013136 APA62634.1 APA62635.1 KK735695 KFR15332.1 LSYS01005191 OPJ78140.1 KK824125 KFP71799.1 KK448607 KFQ72377.1 JH882006 ELR51402.1 CR861261 CAH93329.1
KQ461196 KPJ06347.1 GEZM01051535 JAV74839.1 GEZM01051533 JAV74841.1 KQ971321 EFA00011.2 KYB28740.1 NEVH01020333 PNF21813.1 PNF21812.1 KK852981 KDR12950.1 LJIG01002956 KRT83984.1 KZ288305 PBC28785.1 JH431429 NNAY01004010 OXU18499.1 JH431841 CM004472 OCT84338.1 BC079695 AAH79695.1 OCT84339.1 KQ434809 KZC06556.1 BC121290 AAI21291.1 AEYP01035253 AEYP01035254 KK940306 KFQ50194.1 GBYB01001917 JAG71684.1 AAMC01102941 AAMC01102942 AAMC01102943 AAMC01102944 AAMC01102945 AAMC01102946 AAMC01102947 GL193222 EFB21511.1 JW861845 AFO94362.1 BC123412 AAI23413.1 QUSF01000023 RLW01270.1 KL887256 KGL74715.1 KL669222 KFW75862.1 D42053 AK291773 AC040169 BC114555 BC114961 AL133583 AAEX03004026 AAEX03004027 GEBF01001246 JAO02387.1 KE161800 EPQ05449.1 KB743829 EOA96927.1 KL870139 KGL86881.1 AANG04002512 JP013074 AES01672.1 AKHW03005795 KYO25431.1 AGTP01083736 AGTP01083737 AGTP01083738 ADFV01071441 ADFV01071442 ADFV01071443 AEMK02000039 DQIR01157021 DQIR01213789 DQIR01298853 HDB12498.1 AAKN02054945 AQIA01043817 AQIA01043818 AQIA01043819 AQIA01043820 AQIB01129880 AHZZ02019146 AHZZ02019147 JU477639 AFH34443.1 JU336898 JV048573 AFE80651.1 AFI38644.1 JSUE03026761 JSUE03026762 CM001272 EHH31903.1 GEDV01011589 JAP76968.1 KL447421 KFO73387.1 ABQF01035918 AACZ04051371 AC195663 GABC01003307 GABF01005018 GABD01002378 GABE01010437 NBAG03000295 JAA08031.1 JAA17127.1 JAA30722.1 JAA34302.1 PNI45721.1 DS235430 EEB15675.1 KZ505864 PKU43898.1 KL378584 KFP85686.1 BX548157 CT025908 CAN88262.1 KX013132 KX013134 KX013138 KX013139 KX013140 KX013142 KX013143 KX013144 APA62631.1 APA62633.1 APA62637.1 APA62638.1 APA62639.1 APA62641.1 APA62642.1 APA62643.1 GEDV01008281 JAP80276.1 KL410965 KFR01634.1 KL343431 KFZ54330.1 KX013135 KX013136 APA62634.1 APA62635.1 KK735695 KFR15332.1 LSYS01005191 OPJ78140.1 KK824125 KFP71799.1 KK448607 KFQ72377.1 JH882006 ELR51402.1 CR861261 CAH93329.1
Proteomes
UP000218220
UP000007151
UP000053268
UP000053240
UP000007266
UP000192223
+ More
UP000235965 UP000027135 UP000242457 UP000005203 UP000215335 UP000186698 UP000085678 UP000076502 UP000000715 UP000248482 UP000261680 UP000291021 UP000008143 UP000248481 UP000261681 UP000001646 UP000252040 UP000009136 UP000248483 UP000276834 UP000053641 UP000053258 UP000248480 UP000265300 UP000005640 UP000002254 UP000053858 UP000011712 UP000050525 UP000005215 UP000001073 UP000008227 UP000233060 UP000245320 UP000189705 UP000005447 UP000233120 UP000233100 UP000029965 UP000028761 UP000233140 UP000006718 UP000053760 UP000007754 UP000002277 UP000009046 UP000000437 UP000233180 UP000233200 UP000233080 UP000053283 UP000007646 UP000053605 UP000190648 UP000233020
UP000235965 UP000027135 UP000242457 UP000005203 UP000215335 UP000186698 UP000085678 UP000076502 UP000000715 UP000248482 UP000261680 UP000291021 UP000008143 UP000248481 UP000261681 UP000001646 UP000252040 UP000009136 UP000248483 UP000276834 UP000053641 UP000053258 UP000248480 UP000265300 UP000005640 UP000002254 UP000053858 UP000011712 UP000050525 UP000005215 UP000001073 UP000008227 UP000233060 UP000245320 UP000189705 UP000005447 UP000233120 UP000233100 UP000029965 UP000028761 UP000233140 UP000006718 UP000053760 UP000007754 UP000002277 UP000009046 UP000000437 UP000233180 UP000233200 UP000233080 UP000053283 UP000007646 UP000053605 UP000190648 UP000233020
PRIDE
Pfam
PF00082 Peptidase_S8
Interpro
SUPFAM
SSF52743
SSF52743
Gene 3D
ProteinModelPortal
A0A2A4JC15
A0A212EGG8
A0A194PGX1
A0A194QLD6
A0A1Y1LMH9
A0A1Y1LPH7
+ More
D6WIB2 A0A139WLB7 A0A1W4WJX8 A0A1W4WIY9 A0A2J7PZP8 A0A2J7PZR5 A0A067QSV2 A0A0T6B9G1 A0A2A3EAP1 T1ISC2 A0A088A5Z3 A0A232EJF7 T1J4A8 A0A1L8GKG1 Q6AX98 A0A1L8GKI6 A0A1S3HYQ9 A0A154P3R4 Q0VA23 M3YC21 A0A2Y9K244 A0A384BZG3 A0A091S4F9 A0A0C9QMP8 F6S2F2 D2HRI4 A0A2Y9IG02 V9K9C3 A0A2Y9SY48 A0A384A5I7 G1KN46 A0A341AP56 Q08E55 A0A2Y9P8W9 A0A3L8SFL4 A0A2Y9SPV3 A0A2Y9NXL1 A0A099Z1P2 A0A093PUW3 A0A2Y9DNI6 A0A340X0E6 A0A2Y9K2N3 Q14703 F1PER6 A0A0P6JL18 S7MMG9 R0L9K5 A0A099ZYJ0 M3WFB1 G9K9U7 A0A151MLJ3 I3MBB7 G1RJD6 A0A286ZZH4 A0A2K5KZ34 A0A2U4BRP0 A0A1U7RFK3 A0A1U7R8F2 H0V6Z3 A0A2K6CMT2 A0A2K5V1G2 A0A2K5V1H8 A0A0D9S1D8 A0A2U4BRQ6 A0A096NPH3 H9ZES7 A0A2K5Z4Q0 H9G192 F7G9K2 A0A131YEJ8 A0A091GGY9 H0Z8S1 H2QBL9 E0VQL9 A0A2I0UCW7 A0A091N8D2 Q1LWH3 A0A1I9XGB5 A0A2K6K2W1 A0A2K6R346 A0A2K5IJ30 A0A131YQA9 A0A091VER6 A0A094KTH6 A0A1I9XGB3 G3SXF6 A0A091WI36 A0A1V4K102 A0A091N6X0 A0A091TEW2 L8I827 Q5R4I7 A0A2K5F5D8
D6WIB2 A0A139WLB7 A0A1W4WJX8 A0A1W4WIY9 A0A2J7PZP8 A0A2J7PZR5 A0A067QSV2 A0A0T6B9G1 A0A2A3EAP1 T1ISC2 A0A088A5Z3 A0A232EJF7 T1J4A8 A0A1L8GKG1 Q6AX98 A0A1L8GKI6 A0A1S3HYQ9 A0A154P3R4 Q0VA23 M3YC21 A0A2Y9K244 A0A384BZG3 A0A091S4F9 A0A0C9QMP8 F6S2F2 D2HRI4 A0A2Y9IG02 V9K9C3 A0A2Y9SY48 A0A384A5I7 G1KN46 A0A341AP56 Q08E55 A0A2Y9P8W9 A0A3L8SFL4 A0A2Y9SPV3 A0A2Y9NXL1 A0A099Z1P2 A0A093PUW3 A0A2Y9DNI6 A0A340X0E6 A0A2Y9K2N3 Q14703 F1PER6 A0A0P6JL18 S7MMG9 R0L9K5 A0A099ZYJ0 M3WFB1 G9K9U7 A0A151MLJ3 I3MBB7 G1RJD6 A0A286ZZH4 A0A2K5KZ34 A0A2U4BRP0 A0A1U7RFK3 A0A1U7R8F2 H0V6Z3 A0A2K6CMT2 A0A2K5V1G2 A0A2K5V1H8 A0A0D9S1D8 A0A2U4BRQ6 A0A096NPH3 H9ZES7 A0A2K5Z4Q0 H9G192 F7G9K2 A0A131YEJ8 A0A091GGY9 H0Z8S1 H2QBL9 E0VQL9 A0A2I0UCW7 A0A091N8D2 Q1LWH3 A0A1I9XGB5 A0A2K6K2W1 A0A2K6R346 A0A2K5IJ30 A0A131YQA9 A0A091VER6 A0A094KTH6 A0A1I9XGB3 G3SXF6 A0A091WI36 A0A1V4K102 A0A091N6X0 A0A091TEW2 L8I827 Q5R4I7 A0A2K5F5D8
PDB
1BH6
E-value=2.55657e-20,
Score=247
Ontologies
GO
GO:0004252
GO:0016021
GO:0005794
GO:0006606
GO:0007040
GO:0005795
GO:0006629
GO:0001889
GO:0051216
GO:0044267
GO:0034976
GO:0000139
GO:0005789
GO:0030968
GO:0008203
GO:0031293
GO:0043687
GO:0005788
GO:0045540
GO:0036500
GO:0006508
GO:0004175
GO:0004190
GO:0043565
GO:0043401
GO:0003677
GO:0003707
GO:0007169
Topology
Subcellular location
Endoplasmic reticulum membrane
Golgi apparatus membrane
Golgi apparatus membrane
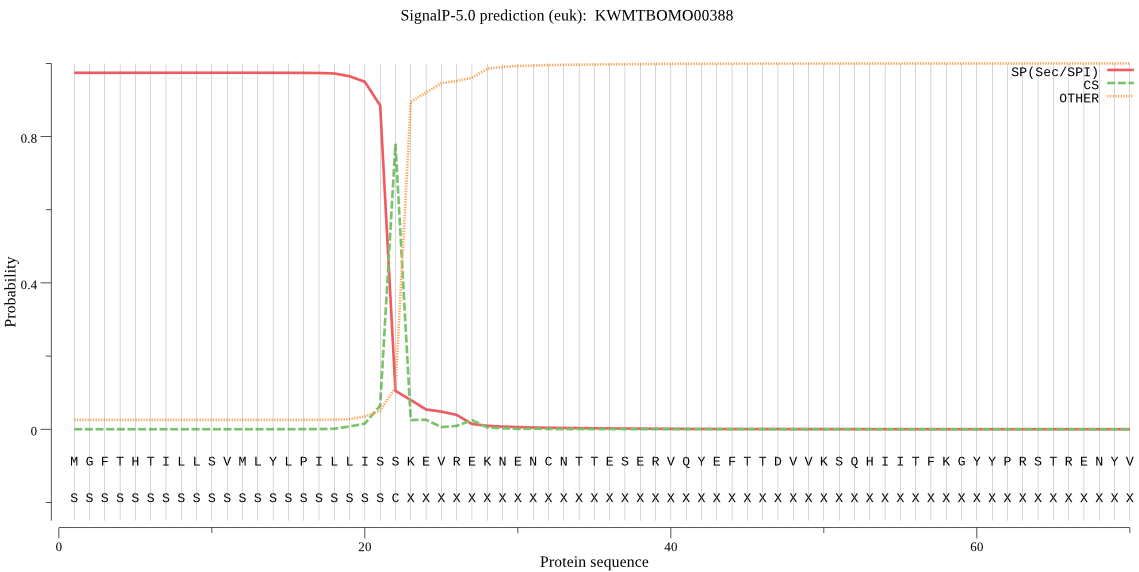
SignalP
Position: 1 - 22,
Likelihood: 0.974286
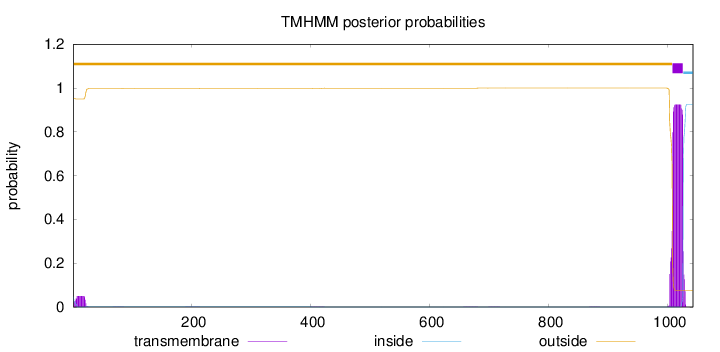
Length:
1042
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
19.1747
Exp number, first 60 AAs:
0.90125
Total prob of N-in:
0.04737
outside
1 - 1007
TMhelix
1008 - 1025
inside
1026 - 1042
Population Genetic Test Statistics
Pi
3.951263
Theta
16.687174
Tajima's D
-2.184838
CLR
0.032736
CSRT
0.00614969251537423
Interpretation
Uncertain
