Gene
KWMTBOMO00387
Pre Gene Modal
BGIBMGA000506
Annotation
PREDICTED:_lysophospholipase-like_protein_1_[Bombyx_mori]
Location in the cell
Mitochondrial Reliability : 2.329
Sequence
CDS
ATGTCTAAGCTAGGAGCTCTTCACATAACCAAGCACAGTGGTGCCAAACAGACCGCTACAGTTATATTTTTTCACGGATCTGGATCCACAGGGGCTGATATCAAAGAATGGGTTAGATTAATGGTTGAACAGTTTTCTTTTCCACATGTGAAAGTTTTGTTTCCGACTGCACCCTTGCAGCCATACACGCCTGCCGGTGGTATGATGAGCAATGTTTGGTTTGATAGAGCTAATATTACACCAGATGTTCCTGAAAAACTGGACTCATTAGCCAGAATTGAAACTGAAGTAAAGAATCTTATAAAAACTGAAAATGATGCTGGAATACCCAGTGATAGGATTATAGTTGGTGGATTTTCCATGGGAGGTGCACTGGCTTTTCACACTGGCTACAGATGGGATAGAAAACTGGCTGGGGTGTTTGCATTCAGCTCCTTCTTAAATTATAATTCAGCCGTTTATGATGAATTGAAAAATAACACAGGTGTAACATATCCACCCTTGCTGCAAATTCATGGTAACCAGGACGATTTAGTACCCCTGAAATGGGGAGAGGAAACATATCAGAAGTTGAAAAGTCTTGGAATTCAGGGAAGTTTTTTCGTTCAAGAAAGATTGGGGCACTCGTTGAATAGACGAGGCATTAAGATTATTAAAGATTGGATCGATGAAAAGTTGCCTAATTCATAA
Protein
MSKLGALHITKHSGAKQTATVIFFHGSGSTGADIKEWVRLMVEQFSFPHVKVLFPTAPLQPYTPAGGMMSNVWFDRANITPDVPEKLDSLARIETEVKNLIKTENDAGIPSDRIIVGGFSMGGALAFHTGYRWDRKLAGVFAFSSFLNYNSAVYDELKNNTGVTYPPLLQIHGNQDDLVPLKWGEETYQKLKSLGIQGSFFVQERLGHSLNRRGIKIIKDWIDEKLPNS
Summary
Uniprot
A0A2H1X2I8
A0A2A4J2A6
A0A0L7KV72
A0A212EGJ8
S4P9Z3
A0A194PGX6
+ More
A0A194QLV0 A0A026WSF5 D2A1N3 A0A1W4VQ45 A0A1B0AVB8 A0A1A9YBM6 A0A1A9VV62 A0A1A9ZM90 A0A1B0GLV9 A0A1L8DYZ4 A0A154PH82 A0A1L8DYY6 B3P1J1 A0A1I8NX81 A0A0L0CTQ4 A0A0M8ZT41 B4QUN3 A0A0J7L6M1 U5EXF5 A0A1W4WU75 E2BV86 A0A1Y1KSP2 B4PLE3 E2ALU6 B4HIN9 Q9VGV9 Q297H5 B4M5I9 R4GHY3 A0A1A9WTW9 A0A2M4BXT6 A0A2M4BYG5 A0A093PFJ3 A0A2M4BXM0 A0A1V4JKI5 A0A2M4BXS4 A0A0A1WVP4 A0A084WP10 B3M2A0 A0A1B6GQS3 A0A2M4AY32 A0A1Q3FHY1 A0A226MII0 U3KCI8 A0A1I8N9S9 A0A0C9QUK2 A0A218U8V2 A0A093GJU3 A0A195B6U6 H0YX80 A0A1S3K9W1 A0A195CP11 E9IBB3 B4JHL7 A0A3B4C1E0 A0A195EG05 A0A158NI38 A0A182FPF8 A0A182QU39 V9L8X7 W5JK72 V4AC82 A0A2A3EQB6 B0W994 B4KHE1 A0A098LZ11 A0A2I0USC1 A0A1B6KUS9 A0A182XX28 A0A3B4A141 A0A2D0S2D6 A0A182KBH5 A0A1A9WTX2 B4NJZ1 A0A088A0H1 B4KC91 A0A182PS64 A0A0M4EGX3 A0A182MI95 A0A182RX74 A0A182JC03 A0A3P8YZI4 A0A182VP10 A0A182L6Z5 A0A182XG11 Q7QAZ5 A0A182ID57 A0A182U7S1 N6TPV0 A0A182W169 A0A210QNP8 A0A091RPI4 A0A0Q3MF39
A0A194QLV0 A0A026WSF5 D2A1N3 A0A1W4VQ45 A0A1B0AVB8 A0A1A9YBM6 A0A1A9VV62 A0A1A9ZM90 A0A1B0GLV9 A0A1L8DYZ4 A0A154PH82 A0A1L8DYY6 B3P1J1 A0A1I8NX81 A0A0L0CTQ4 A0A0M8ZT41 B4QUN3 A0A0J7L6M1 U5EXF5 A0A1W4WU75 E2BV86 A0A1Y1KSP2 B4PLE3 E2ALU6 B4HIN9 Q9VGV9 Q297H5 B4M5I9 R4GHY3 A0A1A9WTW9 A0A2M4BXT6 A0A2M4BYG5 A0A093PFJ3 A0A2M4BXM0 A0A1V4JKI5 A0A2M4BXS4 A0A0A1WVP4 A0A084WP10 B3M2A0 A0A1B6GQS3 A0A2M4AY32 A0A1Q3FHY1 A0A226MII0 U3KCI8 A0A1I8N9S9 A0A0C9QUK2 A0A218U8V2 A0A093GJU3 A0A195B6U6 H0YX80 A0A1S3K9W1 A0A195CP11 E9IBB3 B4JHL7 A0A3B4C1E0 A0A195EG05 A0A158NI38 A0A182FPF8 A0A182QU39 V9L8X7 W5JK72 V4AC82 A0A2A3EQB6 B0W994 B4KHE1 A0A098LZ11 A0A2I0USC1 A0A1B6KUS9 A0A182XX28 A0A3B4A141 A0A2D0S2D6 A0A182KBH5 A0A1A9WTX2 B4NJZ1 A0A088A0H1 B4KC91 A0A182PS64 A0A0M4EGX3 A0A182MI95 A0A182RX74 A0A182JC03 A0A3P8YZI4 A0A182VP10 A0A182L6Z5 A0A182XG11 Q7QAZ5 A0A182ID57 A0A182U7S1 N6TPV0 A0A182W169 A0A210QNP8 A0A091RPI4 A0A0Q3MF39
Pubmed
26227816
22118469
23622113
26354079
24508170
18362917
+ More
19820115 17994087 26108605 20798317 28004739 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 15592404 25830018 24438588 18057021 25315136 20360741 21282665 21347285 24402279 20920257 23761445 23254933 25244985 25463417 25069045 20966253 12364791 14747013 17210077 23537049 28812685
19820115 17994087 26108605 20798317 28004739 17550304 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 15632085 15592404 25830018 24438588 18057021 25315136 20360741 21282665 21347285 24402279 20920257 23761445 23254933 25244985 25463417 25069045 20966253 12364791 14747013 17210077 23537049 28812685
EMBL
ODYU01012939
SOQ59487.1
NWSH01004012
PCG65570.1
JTDY01005292
KOB67147.1
+ More
AGBW02015069 OWR40609.1 GAIX01003489 JAA89071.1 KQ459605 KPI91959.1 KQ461196 KPJ06339.1 KK107111 EZA58962.1 KQ971338 EFA02699.1 JXJN01004198 AJVK01009127 GFDF01002393 JAV11691.1 KQ434905 KZC11221.1 GFDF01002438 JAV11646.1 CH954181 EDV49590.1 JRES01000032 KNC34769.1 KQ435863 KOX70407.1 CM000364 EDX13440.1 LBMM01000451 KMQ98311.1 GANO01002563 JAB57308.1 GL450824 EFN80378.1 GEZM01074909 JAV64419.1 CM000160 EDW96850.1 GL440629 EFN65616.1 CH480815 EDW42686.1 AE014297 AY102661 AAF54564.1 AAM27490.1 CM000070 EAL28230.1 CH940652 EDW58915.1 AADN05000005 GGFJ01008683 MBW57824.1 GGFJ01008820 MBW57961.1 KL669024 KFW75176.1 GGFJ01008684 MBW57825.1 LSYS01007194 OPJ72267.1 GGFJ01008682 MBW57823.1 GBXI01011601 JAD02691.1 ATLV01024808 KE525361 KFB51954.1 CH902617 EDV42291.1 KPU79644.1 GECZ01005066 JAS64703.1 GGFK01012376 MBW45697.1 GFDL01007869 JAV27176.1 MCFN01000801 OXB55097.1 AGTO01020506 GBYB01004312 JAG74079.1 MUZQ01000652 OWK49950.1 KL215855 KFV67059.1 KQ976579 KYM79987.1 ABQF01123408 KQ977481 KYN02451.1 GL762111 EFZ22050.1 CH916369 EDV93856.1 KQ978957 KYN27096.1 ADTU01016263 AXCN02001559 JW875930 AFP08447.1 ADMH02001267 ETN63290.1 KB200454 ESP01609.1 KZ288194 PBC33897.1 DS231862 EDS39791.1 CH933807 EDW11205.1 GBSI01001300 JAC95196.1 KZ505644 PKU48944.1 GEBQ01024780 JAT15197.1 CH964272 EDW83993.1 CH933806 EDW13700.1 CP012526 ALC47704.1 AXCM01000384 AAAB01008880 EAA08531.3 APCN01005034 APGK01057068 KB741277 KB631617 ENN71275.1 ERL84704.1 NEDP02002617 OWF50351.1 KK803361 KFQ30362.1 LMAW01002459 KQK81215.1
AGBW02015069 OWR40609.1 GAIX01003489 JAA89071.1 KQ459605 KPI91959.1 KQ461196 KPJ06339.1 KK107111 EZA58962.1 KQ971338 EFA02699.1 JXJN01004198 AJVK01009127 GFDF01002393 JAV11691.1 KQ434905 KZC11221.1 GFDF01002438 JAV11646.1 CH954181 EDV49590.1 JRES01000032 KNC34769.1 KQ435863 KOX70407.1 CM000364 EDX13440.1 LBMM01000451 KMQ98311.1 GANO01002563 JAB57308.1 GL450824 EFN80378.1 GEZM01074909 JAV64419.1 CM000160 EDW96850.1 GL440629 EFN65616.1 CH480815 EDW42686.1 AE014297 AY102661 AAF54564.1 AAM27490.1 CM000070 EAL28230.1 CH940652 EDW58915.1 AADN05000005 GGFJ01008683 MBW57824.1 GGFJ01008820 MBW57961.1 KL669024 KFW75176.1 GGFJ01008684 MBW57825.1 LSYS01007194 OPJ72267.1 GGFJ01008682 MBW57823.1 GBXI01011601 JAD02691.1 ATLV01024808 KE525361 KFB51954.1 CH902617 EDV42291.1 KPU79644.1 GECZ01005066 JAS64703.1 GGFK01012376 MBW45697.1 GFDL01007869 JAV27176.1 MCFN01000801 OXB55097.1 AGTO01020506 GBYB01004312 JAG74079.1 MUZQ01000652 OWK49950.1 KL215855 KFV67059.1 KQ976579 KYM79987.1 ABQF01123408 KQ977481 KYN02451.1 GL762111 EFZ22050.1 CH916369 EDV93856.1 KQ978957 KYN27096.1 ADTU01016263 AXCN02001559 JW875930 AFP08447.1 ADMH02001267 ETN63290.1 KB200454 ESP01609.1 KZ288194 PBC33897.1 DS231862 EDS39791.1 CH933807 EDW11205.1 GBSI01001300 JAC95196.1 KZ505644 PKU48944.1 GEBQ01024780 JAT15197.1 CH964272 EDW83993.1 CH933806 EDW13700.1 CP012526 ALC47704.1 AXCM01000384 AAAB01008880 EAA08531.3 APCN01005034 APGK01057068 KB741277 KB631617 ENN71275.1 ERL84704.1 NEDP02002617 OWF50351.1 KK803361 KFQ30362.1 LMAW01002459 KQK81215.1
Proteomes
UP000218220
UP000037510
UP000007151
UP000053268
UP000053240
UP000053097
+ More
UP000007266 UP000192221 UP000092460 UP000092443 UP000078200 UP000092445 UP000092462 UP000076502 UP000008711 UP000095300 UP000037069 UP000053105 UP000000304 UP000036403 UP000192223 UP000008237 UP000002282 UP000000311 UP000001292 UP000000803 UP000001819 UP000008792 UP000000539 UP000091820 UP000053258 UP000190648 UP000030765 UP000007801 UP000198323 UP000016665 UP000095301 UP000197619 UP000053875 UP000078540 UP000007754 UP000085678 UP000078542 UP000001070 UP000261440 UP000078492 UP000005205 UP000069272 UP000075886 UP000000673 UP000030746 UP000242457 UP000002320 UP000009192 UP000076408 UP000261520 UP000221080 UP000075881 UP000007798 UP000005203 UP000075885 UP000092553 UP000075883 UP000075900 UP000075880 UP000265140 UP000075903 UP000075882 UP000076407 UP000007062 UP000075840 UP000075902 UP000019118 UP000030742 UP000075920 UP000242188 UP000051836
UP000007266 UP000192221 UP000092460 UP000092443 UP000078200 UP000092445 UP000092462 UP000076502 UP000008711 UP000095300 UP000037069 UP000053105 UP000000304 UP000036403 UP000192223 UP000008237 UP000002282 UP000000311 UP000001292 UP000000803 UP000001819 UP000008792 UP000000539 UP000091820 UP000053258 UP000190648 UP000030765 UP000007801 UP000198323 UP000016665 UP000095301 UP000197619 UP000053875 UP000078540 UP000007754 UP000085678 UP000078542 UP000001070 UP000261440 UP000078492 UP000005205 UP000069272 UP000075886 UP000000673 UP000030746 UP000242457 UP000002320 UP000009192 UP000076408 UP000261520 UP000221080 UP000075881 UP000007798 UP000005203 UP000075885 UP000092553 UP000075883 UP000075900 UP000075880 UP000265140 UP000075903 UP000075882 UP000076407 UP000007062 UP000075840 UP000075902 UP000019118 UP000030742 UP000075920 UP000242188 UP000051836
PRIDE
Pfam
PF02230 Abhydrolase_2
SUPFAM
SSF53474
SSF53474
Gene 3D
ProteinModelPortal
A0A2H1X2I8
A0A2A4J2A6
A0A0L7KV72
A0A212EGJ8
S4P9Z3
A0A194PGX6
+ More
A0A194QLV0 A0A026WSF5 D2A1N3 A0A1W4VQ45 A0A1B0AVB8 A0A1A9YBM6 A0A1A9VV62 A0A1A9ZM90 A0A1B0GLV9 A0A1L8DYZ4 A0A154PH82 A0A1L8DYY6 B3P1J1 A0A1I8NX81 A0A0L0CTQ4 A0A0M8ZT41 B4QUN3 A0A0J7L6M1 U5EXF5 A0A1W4WU75 E2BV86 A0A1Y1KSP2 B4PLE3 E2ALU6 B4HIN9 Q9VGV9 Q297H5 B4M5I9 R4GHY3 A0A1A9WTW9 A0A2M4BXT6 A0A2M4BYG5 A0A093PFJ3 A0A2M4BXM0 A0A1V4JKI5 A0A2M4BXS4 A0A0A1WVP4 A0A084WP10 B3M2A0 A0A1B6GQS3 A0A2M4AY32 A0A1Q3FHY1 A0A226MII0 U3KCI8 A0A1I8N9S9 A0A0C9QUK2 A0A218U8V2 A0A093GJU3 A0A195B6U6 H0YX80 A0A1S3K9W1 A0A195CP11 E9IBB3 B4JHL7 A0A3B4C1E0 A0A195EG05 A0A158NI38 A0A182FPF8 A0A182QU39 V9L8X7 W5JK72 V4AC82 A0A2A3EQB6 B0W994 B4KHE1 A0A098LZ11 A0A2I0USC1 A0A1B6KUS9 A0A182XX28 A0A3B4A141 A0A2D0S2D6 A0A182KBH5 A0A1A9WTX2 B4NJZ1 A0A088A0H1 B4KC91 A0A182PS64 A0A0M4EGX3 A0A182MI95 A0A182RX74 A0A182JC03 A0A3P8YZI4 A0A182VP10 A0A182L6Z5 A0A182XG11 Q7QAZ5 A0A182ID57 A0A182U7S1 N6TPV0 A0A182W169 A0A210QNP8 A0A091RPI4 A0A0Q3MF39
A0A194QLV0 A0A026WSF5 D2A1N3 A0A1W4VQ45 A0A1B0AVB8 A0A1A9YBM6 A0A1A9VV62 A0A1A9ZM90 A0A1B0GLV9 A0A1L8DYZ4 A0A154PH82 A0A1L8DYY6 B3P1J1 A0A1I8NX81 A0A0L0CTQ4 A0A0M8ZT41 B4QUN3 A0A0J7L6M1 U5EXF5 A0A1W4WU75 E2BV86 A0A1Y1KSP2 B4PLE3 E2ALU6 B4HIN9 Q9VGV9 Q297H5 B4M5I9 R4GHY3 A0A1A9WTW9 A0A2M4BXT6 A0A2M4BYG5 A0A093PFJ3 A0A2M4BXM0 A0A1V4JKI5 A0A2M4BXS4 A0A0A1WVP4 A0A084WP10 B3M2A0 A0A1B6GQS3 A0A2M4AY32 A0A1Q3FHY1 A0A226MII0 U3KCI8 A0A1I8N9S9 A0A0C9QUK2 A0A218U8V2 A0A093GJU3 A0A195B6U6 H0YX80 A0A1S3K9W1 A0A195CP11 E9IBB3 B4JHL7 A0A3B4C1E0 A0A195EG05 A0A158NI38 A0A182FPF8 A0A182QU39 V9L8X7 W5JK72 V4AC82 A0A2A3EQB6 B0W994 B4KHE1 A0A098LZ11 A0A2I0USC1 A0A1B6KUS9 A0A182XX28 A0A3B4A141 A0A2D0S2D6 A0A182KBH5 A0A1A9WTX2 B4NJZ1 A0A088A0H1 B4KC91 A0A182PS64 A0A0M4EGX3 A0A182MI95 A0A182RX74 A0A182JC03 A0A3P8YZI4 A0A182VP10 A0A182L6Z5 A0A182XG11 Q7QAZ5 A0A182ID57 A0A182U7S1 N6TPV0 A0A182W169 A0A210QNP8 A0A091RPI4 A0A0Q3MF39
PDB
5KRE
E-value=4.75645e-50,
Score=497
Ontologies
GO
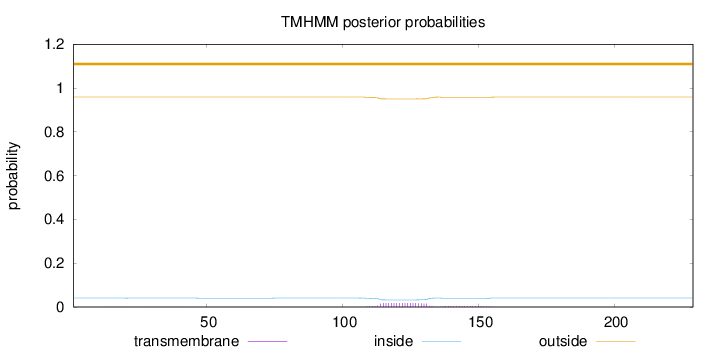
Topology
Length:
229
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.43179
Exp number, first 60 AAs:
0.01825
Total prob of N-in:
0.04117
outside
1 - 229
Population Genetic Test Statistics
Pi
5.029719
Theta
18.145349
Tajima's D
-1.459908
CLR
0.376892
CSRT
0.0633468326583671
Interpretation
Uncertain
