Gene
KWMTBOMO00375 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000715
Annotation
probable_medium-chain_specific_acyl-CoA_dehydrogenase?_mitochondrial_[Papilio_xuthus]
Full name
Probable medium-chain specific acyl-CoA dehydrogenase, mitochondrial
Location in the cell
Mitochondrial Reliability : 2.485
Sequence
CDS
ATGAACCCGTTGGCGCAGATTGTTCGTGCCACTCGCCCTATTTACAGGAAATTATCAACAACTGCAACAACTGCTTCTGCTGCAAAGCCAATACCGACAACAGGCTTTTGTTTCGAATTAACCGATGAGCAGAAAGCGCTTCAGGAACTAGCGCGTAAATTCACGAAGGATGAAATCATTCCTGTGGCGGGTCAATATGATAAGACGGGGGAGTACCCGTGGCCCGTTGTAAAGAAGGCTTGGGAAGTTGGCCTAATGAACGGACACATTCCAGAACATTGCGGGGGCATAGGGAATTTTGGCGTCTTCGAAGAGTGCATCGTAGCGGACGAACTTGCGTTTGGATGTACTGGCATCACGACAGCTATTGGGGGCACGTCCTTAGGACAAACTCCCATCATTATTGCTGGAAATAAGGAGCAGCAGAAGAAATATCTGGGTAGACTAATCGACGAACCATTAGTCGCTGCCTATGGGGTGACCGAACCTGGTGCTGGATCCGATGTAGCCGGTATAAAGACGAGAGCGGAGAAGAAAGGCGACGAATGGATCCTGAACGGTCAGAAAATGTGGATCACCAACGGTGGTGTTGCCAACTGGTACTTTGTATTGGCTCGTACGAATCCGGACCCTAAATGTCCGGCCAGCAAAGCATTCACCGGCTTCATCGTCGAACGGGAATGGCCTGGGGTCACTCCAGGGCGAAAGGAACAAAACATGGGTCAGCGTGCTTCGGACACCCGAGGTATCACGTTTGAAGATGTGCGTATCCCCAAAGAAAATGTGCTTATTGGCGAAGGAGCTGGATTCAAGATCGCCATGGGAGCCTTTGACAAGACTCGTCCTCCGGTAGCGGCTGGTGCTACTGGGTTGGCTCAACGGGCTCTCCACGAAGCCACCAAGTACTCGTTGGAACGCAAAACGTTCGGAGTGCCCATAGCGAGACACCAGGCGGTAGCGTTCATGCTTGCCGACATGGCGATAGGAGTGGAGACCGCTCGTATCGCATGGCAACGCGCCGCATGGATGGTTGACCACGGACAAAAGAATACAGTTATGGCGTCAGTGGCCAAGTGTCACGCTTCGGAAATCGCTAACAAGGCAGCCACTGATGCGGTCCAAGTGTTCGGAGGAAACGGCTTCAACACCGAATACCCAGTCGAGAAACTGATGCGGGACGCCAAGATATACCAAATCTACGAAGGAACCTCGCAGATTCAAAGGCTCATCATATCAAGAGAGATCCTCACCGCAGCAACTCAAACCAACTGA
Protein
MNPLAQIVRATRPIYRKLSTTATTASAAKPIPTTGFCFELTDEQKALQELARKFTKDEIIPVAGQYDKTGEYPWPVVKKAWEVGLMNGHIPEHCGGIGNFGVFEECIVADELAFGCTGITTAIGGTSLGQTPIIIAGNKEQQKKYLGRLIDEPLVAAYGVTEPGAGSDVAGIKTRAEKKGDEWILNGQKMWITNGGVANWYFVLARTNPDPKCPASKAFTGFIVEREWPGVTPGRKEQNMGQRASDTRGITFEDVRIPKENVLIGEGAGFKIAMGAFDKTRPPVAAGATGLAQRALHEATKYSLERKTFGVPIARHQAVAFMLADMAIGVETARIAWQRAAWMVDHGQKNTVMASVAKCHASEIANKAATDAVQVFGGNGFNTEYPVEKLMRDAKIYQIYEGTSQIQRLIISREILTAATQTN
Summary
Description
This enzyme is specific for acyl chain lengths of 4 to 16.
Catalytic Activity
a medium-chain 2,3-saturated fatty acyl-CoA + H(+) + oxidized [electron-transfer flavoprotein] = a medium-chain trans-(2E)-enoyl-CoA + reduced [electron-transfer flavoprotein]
Cofactor
FAD
Subunit
Homotetramer.
Similarity
Belongs to the acyl-CoA dehydrogenase family.
Keywords
Complete proteome
FAD
Fatty acid metabolism
Flavoprotein
Lipid metabolism
Mitochondrion
Oxidoreductase
Reference proteome
Transit peptide
Feature
chain Probable medium-chain specific acyl-CoA dehydrogenase, mitochondrial
Uniprot
I4DJ33
A0A194PLJ7
A0A212EGL0
A0A068FRF0
D2XMK9
H9ITY5
+ More
A0A194QMX3 A0A0M4E8L7 B3M619 B4N6W6 A0A1W4WYU1 A0A3B0KHR1 Q2M113 B4GQT6 B4PCA8 A0A1W4VNC1 B3NF95 V5G253 V5I8N3 A0A182WIG5 B4IX89 G3KKP7 Q9VSA3 B4L0Z8 B4LGR7 A0A0J9RR61 B4HIV1 A0A182KGT3 A0A1I8MQB6 A0A2J7PW45 A0A2J7PW54 A0A2J7PW50 A0A182PHN8 A0A182Q9A6 A0A182MS61 A0A2J7PW57 Q16GA1 B4QKX0 A0A0L0CHL3 A0A1L8EE47 A0A182UV67 A0A182XMV3 A0A182LJ14 Q7PNP8 A0A182HWQ9 A0A182UAW2 A0A182RFA8 A0A1I8NNJ1 A0A023ERV4 A0A182Y1B6 A0A182NGY1 A0A1Q3FJJ8 A0A023F9Y8 A0A067QLW6 A0A182GQZ1 D6WFZ1 A0A182IJX2 A0A1A9WG17 A0A084W486 B0WLI6 A0A2R7WI88 A0A1A9UPF3 A0A034WER3 A0A1I9WLC3 A0A1A9XCG3 A0A1B0B8Q9 A0A3B0JW24 A0A2M4A5A9 A0A067QSV4 Q1HQM4 T1E7Y7 A0A1A9ZBP0 A0A182FH31 A0A069DUA7 A0A0K8W6D8 D3TNV6 A0A1L8E2A5 U5EWS3 W8BI95 T1PAW1 A0A2M4CT55 A0A2M4A6I5 A0A2M4BP80 A0A0P4VYB9 A0A2M4BPB6 R4G3Z8 A0A2M3ZFS0 B5DG29 A0A2M4A7U1 A0A1Y1MQH3 A0A0A1X935 A0A1L8E2S6 A0A2J7QMR2 A0A2M3Z061 A0A060YMD9 A0A0A9ZCR3 A0A0K8TTW8 A0A1B6GZ82 D6WEK6 A0A0P5FQZ8
A0A194QMX3 A0A0M4E8L7 B3M619 B4N6W6 A0A1W4WYU1 A0A3B0KHR1 Q2M113 B4GQT6 B4PCA8 A0A1W4VNC1 B3NF95 V5G253 V5I8N3 A0A182WIG5 B4IX89 G3KKP7 Q9VSA3 B4L0Z8 B4LGR7 A0A0J9RR61 B4HIV1 A0A182KGT3 A0A1I8MQB6 A0A2J7PW45 A0A2J7PW54 A0A2J7PW50 A0A182PHN8 A0A182Q9A6 A0A182MS61 A0A2J7PW57 Q16GA1 B4QKX0 A0A0L0CHL3 A0A1L8EE47 A0A182UV67 A0A182XMV3 A0A182LJ14 Q7PNP8 A0A182HWQ9 A0A182UAW2 A0A182RFA8 A0A1I8NNJ1 A0A023ERV4 A0A182Y1B6 A0A182NGY1 A0A1Q3FJJ8 A0A023F9Y8 A0A067QLW6 A0A182GQZ1 D6WFZ1 A0A182IJX2 A0A1A9WG17 A0A084W486 B0WLI6 A0A2R7WI88 A0A1A9UPF3 A0A034WER3 A0A1I9WLC3 A0A1A9XCG3 A0A1B0B8Q9 A0A3B0JW24 A0A2M4A5A9 A0A067QSV4 Q1HQM4 T1E7Y7 A0A1A9ZBP0 A0A182FH31 A0A069DUA7 A0A0K8W6D8 D3TNV6 A0A1L8E2A5 U5EWS3 W8BI95 T1PAW1 A0A2M4CT55 A0A2M4A6I5 A0A2M4BP80 A0A0P4VYB9 A0A2M4BPB6 R4G3Z8 A0A2M3ZFS0 B5DG29 A0A2M4A7U1 A0A1Y1MQH3 A0A0A1X935 A0A1L8E2S6 A0A2J7QMR2 A0A2M3Z061 A0A060YMD9 A0A0A9ZCR3 A0A0K8TTW8 A0A1B6GZ82 D6WEK6 A0A0P5FQZ8
EC Number
1.3.8.7
Pubmed
22651552
26354079
22118469
26385554
20074338
19121390
+ More
17994087 15632085 17550304 10731132 12537572 12537569 18057021 22936249 25315136 17510324 26108605 20966253 12364791 14747013 17210077 24945155 26483478 25244985 25474469 24845553 18362917 19820115 24438588 25348373 27538518 17204158 26334808 20353571 24495485 27129103 19878547 20433749 28004739 25830018 24755649 25401762 26823975 26369729
17994087 15632085 17550304 10731132 12537572 12537569 18057021 22936249 25315136 17510324 26108605 20966253 12364791 14747013 17210077 24945155 26483478 25244985 25474469 24845553 18362917 19820115 24438588 25348373 27538518 17204158 26334808 20353571 24495485 27129103 19878547 20433749 28004739 25830018 24755649 25401762 26823975 26369729
EMBL
AK401301
BAM17923.1
KQ459605
KPI91980.1
AGBW02015069
OWR40617.1
+ More
KJ622080 AID66670.1 GU205155 ADB57042.1 BABH01007168 BABH01007169 KQ461196 KPJ06320.1 CP012525 ALC43241.1 CH902618 EDV40735.1 CH964168 EDW80105.1 OUUW01000009 SPP84641.1 CH379069 EAL30764.1 CH479188 EDW40121.1 CM000159 EDW93793.1 CH954178 EDV50437.1 GALX01004386 JAB64080.1 GALX01004385 JAB64081.1 CH916366 EDV97421.1 JN252151 JN252152 JN252153 JN252154 JN252155 JN252156 JN252157 JN252158 JN252159 JN252160 JN252161 JN252162 JN252163 JN252164 JN252165 JN252166 JN252167 JN252168 JN252169 JN252170 JN252171 JN252172 AEO11957.1 AEO11958.1 AEO11959.1 AEO11960.1 AEO11961.1 AEO11962.1 AEO11963.1 AEO11964.1 AEO11965.1 AEO11966.1 AEO11967.1 AEO11968.1 AEO11969.1 AEO11970.1 AEO11971.1 AEO11972.1 AEO11973.1 AEO11974.1 AEO11975.1 AEO11976.1 AEO11977.1 AEO11978.1 AE014296 AY089546 CH933809 EDW18155.1 CH941131 CH940647 EDW57380.1 EDW70532.1 CM002912 KMY98197.1 CH480815 EDW40605.1 NEVH01020938 PNF20562.1 PNF20564.1 PNF20565.1 AXCN02000356 AXCM01017076 PNF20561.1 CH478303 EAT33262.1 CM000363 EDX09603.1 JRES01000375 KNC31868.1 GFDG01001812 JAV16987.1 AAAB01008960 EAA11811.3 APCN01001526 JXUM01094639 JXUM01094640 JXUM01094641 GAPW01001641 KQ564143 JAC11957.1 KXJ72689.1 GFDL01007288 JAV27757.1 GBBI01000898 JAC17814.1 KK853183 KDR10129.1 JXUM01081493 JXUM01081494 JXUM01081495 KQ563250 KXJ74210.1 KQ971328 EEZ99771.1 ATLV01020289 KE525297 KFB45030.1 DS231986 EDS30505.1 KK854849 PTY19219.1 GAKP01006155 JAC52797.1 KU932307 APA33943.1 JXJN01010041 SPP84642.1 GGFK01002675 MBW35996.1 KK853058 KDR11928.1 DQ440420 ABF18453.1 GAMD01003228 JAA98362.1 GBGD01001597 JAC87292.1 GDHF01005623 JAI46691.1 CCAG010020781 EZ423108 ADD19384.1 GFDF01001211 JAV12873.1 GANO01001282 JAB58589.1 GAMC01005590 JAC00966.1 KA645068 AFP59697.1 GGFL01004358 MBW68536.1 GGFK01003092 MBW36413.1 GGFJ01005681 MBW54822.1 GDKW01002026 JAI54569.1 GGFJ01005682 MBW54823.1 ACPB03023307 GAHY01001780 JAA75730.1 GGFM01006655 MBW27406.1 BT043588 BT059085 ACH70703.1 ACN10798.1 GGFK01003387 MBW36708.1 GEZM01027474 JAV86725.1 GBXI01006881 JAD07411.1 GFDF01001212 JAV12872.1 NEVH01013204 PNF29880.1 GGFM01001125 MBW21876.1 FR914393 CDQ93058.1 GBHO01021106 GBHO01021081 GBHO01020140 GBHO01011724 GBHO01011716 GBHO01001910 GBHO01001909 GBHO01001908 GBHO01001905 GBHO01001902 GBRD01015497 GBRD01015496 GBRD01015495 GBRD01015494 GBRD01015493 GBRD01015492 GBRD01015491 GBRD01005481 GBRD01005477 GBRD01005476 GBRD01005475 GBRD01005474 GBRD01005473 GBRD01005472 GBRD01000743 GDHC01019813 JAG22498.1 JAG22523.1 JAG23464.1 JAG31880.1 JAG31888.1 JAG41694.1 JAG41695.1 JAG41696.1 JAG41699.1 JAG41702.1 JAG50329.1 JAP98815.1 GDAI01000208 JAI17395.1 GECZ01002038 JAS67731.1 KQ971318 EFA00426.1 GDIQ01253302 GDIP01140198 JAJ98422.1 JAL63516.1
KJ622080 AID66670.1 GU205155 ADB57042.1 BABH01007168 BABH01007169 KQ461196 KPJ06320.1 CP012525 ALC43241.1 CH902618 EDV40735.1 CH964168 EDW80105.1 OUUW01000009 SPP84641.1 CH379069 EAL30764.1 CH479188 EDW40121.1 CM000159 EDW93793.1 CH954178 EDV50437.1 GALX01004386 JAB64080.1 GALX01004385 JAB64081.1 CH916366 EDV97421.1 JN252151 JN252152 JN252153 JN252154 JN252155 JN252156 JN252157 JN252158 JN252159 JN252160 JN252161 JN252162 JN252163 JN252164 JN252165 JN252166 JN252167 JN252168 JN252169 JN252170 JN252171 JN252172 AEO11957.1 AEO11958.1 AEO11959.1 AEO11960.1 AEO11961.1 AEO11962.1 AEO11963.1 AEO11964.1 AEO11965.1 AEO11966.1 AEO11967.1 AEO11968.1 AEO11969.1 AEO11970.1 AEO11971.1 AEO11972.1 AEO11973.1 AEO11974.1 AEO11975.1 AEO11976.1 AEO11977.1 AEO11978.1 AE014296 AY089546 CH933809 EDW18155.1 CH941131 CH940647 EDW57380.1 EDW70532.1 CM002912 KMY98197.1 CH480815 EDW40605.1 NEVH01020938 PNF20562.1 PNF20564.1 PNF20565.1 AXCN02000356 AXCM01017076 PNF20561.1 CH478303 EAT33262.1 CM000363 EDX09603.1 JRES01000375 KNC31868.1 GFDG01001812 JAV16987.1 AAAB01008960 EAA11811.3 APCN01001526 JXUM01094639 JXUM01094640 JXUM01094641 GAPW01001641 KQ564143 JAC11957.1 KXJ72689.1 GFDL01007288 JAV27757.1 GBBI01000898 JAC17814.1 KK853183 KDR10129.1 JXUM01081493 JXUM01081494 JXUM01081495 KQ563250 KXJ74210.1 KQ971328 EEZ99771.1 ATLV01020289 KE525297 KFB45030.1 DS231986 EDS30505.1 KK854849 PTY19219.1 GAKP01006155 JAC52797.1 KU932307 APA33943.1 JXJN01010041 SPP84642.1 GGFK01002675 MBW35996.1 KK853058 KDR11928.1 DQ440420 ABF18453.1 GAMD01003228 JAA98362.1 GBGD01001597 JAC87292.1 GDHF01005623 JAI46691.1 CCAG010020781 EZ423108 ADD19384.1 GFDF01001211 JAV12873.1 GANO01001282 JAB58589.1 GAMC01005590 JAC00966.1 KA645068 AFP59697.1 GGFL01004358 MBW68536.1 GGFK01003092 MBW36413.1 GGFJ01005681 MBW54822.1 GDKW01002026 JAI54569.1 GGFJ01005682 MBW54823.1 ACPB03023307 GAHY01001780 JAA75730.1 GGFM01006655 MBW27406.1 BT043588 BT059085 ACH70703.1 ACN10798.1 GGFK01003387 MBW36708.1 GEZM01027474 JAV86725.1 GBXI01006881 JAD07411.1 GFDF01001212 JAV12872.1 NEVH01013204 PNF29880.1 GGFM01001125 MBW21876.1 FR914393 CDQ93058.1 GBHO01021106 GBHO01021081 GBHO01020140 GBHO01011724 GBHO01011716 GBHO01001910 GBHO01001909 GBHO01001908 GBHO01001905 GBHO01001902 GBRD01015497 GBRD01015496 GBRD01015495 GBRD01015494 GBRD01015493 GBRD01015492 GBRD01015491 GBRD01005481 GBRD01005477 GBRD01005476 GBRD01005475 GBRD01005474 GBRD01005473 GBRD01005472 GBRD01000743 GDHC01019813 JAG22498.1 JAG22523.1 JAG23464.1 JAG31880.1 JAG31888.1 JAG41694.1 JAG41695.1 JAG41696.1 JAG41699.1 JAG41702.1 JAG50329.1 JAP98815.1 GDAI01000208 JAI17395.1 GECZ01002038 JAS67731.1 KQ971318 EFA00426.1 GDIQ01253302 GDIP01140198 JAJ98422.1 JAL63516.1
Proteomes
UP000053268
UP000007151
UP000005204
UP000053240
UP000092553
UP000007801
+ More
UP000007798 UP000192223 UP000268350 UP000001819 UP000008744 UP000002282 UP000192221 UP000008711 UP000075920 UP000001070 UP000000803 UP000009192 UP000008792 UP000001292 UP000075881 UP000095301 UP000235965 UP000075885 UP000075886 UP000075883 UP000008820 UP000000304 UP000037069 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000075902 UP000075900 UP000095300 UP000069940 UP000249989 UP000076408 UP000075884 UP000027135 UP000007266 UP000075880 UP000091820 UP000030765 UP000002320 UP000078200 UP000092443 UP000092460 UP000092445 UP000069272 UP000092444 UP000015103 UP000087266 UP000193380
UP000007798 UP000192223 UP000268350 UP000001819 UP000008744 UP000002282 UP000192221 UP000008711 UP000075920 UP000001070 UP000000803 UP000009192 UP000008792 UP000001292 UP000075881 UP000095301 UP000235965 UP000075885 UP000075886 UP000075883 UP000008820 UP000000304 UP000037069 UP000075903 UP000076407 UP000075882 UP000007062 UP000075840 UP000075902 UP000075900 UP000095300 UP000069940 UP000249989 UP000076408 UP000075884 UP000027135 UP000007266 UP000075880 UP000091820 UP000030765 UP000002320 UP000078200 UP000092443 UP000092460 UP000092445 UP000069272 UP000092444 UP000015103 UP000087266 UP000193380
PRIDE
Interpro
Gene 3D
CDD
ProteinModelPortal
I4DJ33
A0A194PLJ7
A0A212EGL0
A0A068FRF0
D2XMK9
H9ITY5
+ More
A0A194QMX3 A0A0M4E8L7 B3M619 B4N6W6 A0A1W4WYU1 A0A3B0KHR1 Q2M113 B4GQT6 B4PCA8 A0A1W4VNC1 B3NF95 V5G253 V5I8N3 A0A182WIG5 B4IX89 G3KKP7 Q9VSA3 B4L0Z8 B4LGR7 A0A0J9RR61 B4HIV1 A0A182KGT3 A0A1I8MQB6 A0A2J7PW45 A0A2J7PW54 A0A2J7PW50 A0A182PHN8 A0A182Q9A6 A0A182MS61 A0A2J7PW57 Q16GA1 B4QKX0 A0A0L0CHL3 A0A1L8EE47 A0A182UV67 A0A182XMV3 A0A182LJ14 Q7PNP8 A0A182HWQ9 A0A182UAW2 A0A182RFA8 A0A1I8NNJ1 A0A023ERV4 A0A182Y1B6 A0A182NGY1 A0A1Q3FJJ8 A0A023F9Y8 A0A067QLW6 A0A182GQZ1 D6WFZ1 A0A182IJX2 A0A1A9WG17 A0A084W486 B0WLI6 A0A2R7WI88 A0A1A9UPF3 A0A034WER3 A0A1I9WLC3 A0A1A9XCG3 A0A1B0B8Q9 A0A3B0JW24 A0A2M4A5A9 A0A067QSV4 Q1HQM4 T1E7Y7 A0A1A9ZBP0 A0A182FH31 A0A069DUA7 A0A0K8W6D8 D3TNV6 A0A1L8E2A5 U5EWS3 W8BI95 T1PAW1 A0A2M4CT55 A0A2M4A6I5 A0A2M4BP80 A0A0P4VYB9 A0A2M4BPB6 R4G3Z8 A0A2M3ZFS0 B5DG29 A0A2M4A7U1 A0A1Y1MQH3 A0A0A1X935 A0A1L8E2S6 A0A2J7QMR2 A0A2M3Z061 A0A060YMD9 A0A0A9ZCR3 A0A0K8TTW8 A0A1B6GZ82 D6WEK6 A0A0P5FQZ8
A0A194QMX3 A0A0M4E8L7 B3M619 B4N6W6 A0A1W4WYU1 A0A3B0KHR1 Q2M113 B4GQT6 B4PCA8 A0A1W4VNC1 B3NF95 V5G253 V5I8N3 A0A182WIG5 B4IX89 G3KKP7 Q9VSA3 B4L0Z8 B4LGR7 A0A0J9RR61 B4HIV1 A0A182KGT3 A0A1I8MQB6 A0A2J7PW45 A0A2J7PW54 A0A2J7PW50 A0A182PHN8 A0A182Q9A6 A0A182MS61 A0A2J7PW57 Q16GA1 B4QKX0 A0A0L0CHL3 A0A1L8EE47 A0A182UV67 A0A182XMV3 A0A182LJ14 Q7PNP8 A0A182HWQ9 A0A182UAW2 A0A182RFA8 A0A1I8NNJ1 A0A023ERV4 A0A182Y1B6 A0A182NGY1 A0A1Q3FJJ8 A0A023F9Y8 A0A067QLW6 A0A182GQZ1 D6WFZ1 A0A182IJX2 A0A1A9WG17 A0A084W486 B0WLI6 A0A2R7WI88 A0A1A9UPF3 A0A034WER3 A0A1I9WLC3 A0A1A9XCG3 A0A1B0B8Q9 A0A3B0JW24 A0A2M4A5A9 A0A067QSV4 Q1HQM4 T1E7Y7 A0A1A9ZBP0 A0A182FH31 A0A069DUA7 A0A0K8W6D8 D3TNV6 A0A1L8E2A5 U5EWS3 W8BI95 T1PAW1 A0A2M4CT55 A0A2M4A6I5 A0A2M4BP80 A0A0P4VYB9 A0A2M4BPB6 R4G3Z8 A0A2M3ZFS0 B5DG29 A0A2M4A7U1 A0A1Y1MQH3 A0A0A1X935 A0A1L8E2S6 A0A2J7QMR2 A0A2M3Z061 A0A060YMD9 A0A0A9ZCR3 A0A0K8TTW8 A0A1B6GZ82 D6WEK6 A0A0P5FQZ8
PDB
2A1T
E-value=4.43854e-161,
Score=1457
Ontologies
PATHWAY
GO
Topology
Subcellular location
Mitochondrion matrix
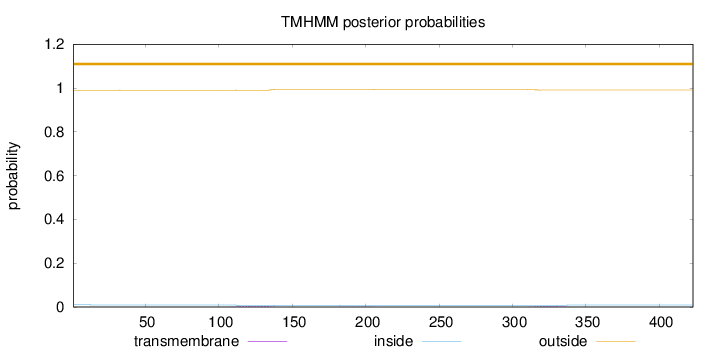
Length:
423
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.13667
Exp number, first 60 AAs:
0.00217
Total prob of N-in:
0.00976
outside
1 - 423
Population Genetic Test Statistics
Pi
16.448881
Theta
20.779237
Tajima's D
-0.89607
CLR
0.150119
CSRT
0.154792260386981
Interpretation
Uncertain
