Gene
KWMTBOMO00373 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000714
Annotation
PREDICTED:_proton-coupled_amino_acid_transporter_4_[Amyelois_transitella]
Location in the cell
PlasmaMembrane Reliability : 4.642
Sequence
CDS
ATGGTGAACGAACCAAATGGCGCCATTCCAGCGCCACAGGAGTTGGAAACATTTATATCTCCGGATGAAAAGAACAAAGAAAAGATTGTTAGCAAATATAACATGACGAAAGATGCGGAGTCAGGAGACTTTGATCCGTTCACCGAAAGGAAGTTGGACACTCCTACTTCGAATATGGACACTCTAACACACTTATTGAAGGCTTCCCTGGGTACTGGAATCTTAGCCATGCCCAAAGCCTTCAAGGCAGCCGGATTGATCAATGGAATACTCTTCACGGTTATCGTGGCCGTCGTCTGCACTCACTGCTCGTATATACTTGTTAAATGTGCCCACGTTTTGTACAAGAAGACGAAAAAAACTCAGATGTCCTTTGCGGAAGTGGGCGAGGCGGCTTTGGATAACGGTCCACAGCCGTTAAGAAAATGGGCGAATGCTTTCAGGATATTCATCGTCATTAGTCTCTTTATAACGTACTTTGGGACGTGCTCCGTCTATACGGTCATAATAGCACGAAACATTTTACAGATAATTCAGTTCTACTGCGGCGATATTATCAATGAACGTTTTCTCATTATAATATTACTGGTCCCACTTATTCTCATGGCCTGGATTCGAAATCTCAAGTACTTAGCTCCTGTGTCAATGATCGCGAATTTATTCATGGCTGTGGGTCTAGGGATCACCCTTTACTACTTAGTAGGTACTGGACAACTGCAATACGATAAGATCAAGGACATGCTCTTCAAACATCCCAGCGAATGGCCCGAATTCTTTTCGCTCACAATTTTCGCAATGGAAGCCATCGGAGTTGTGATGCCTCTCGAAAACTCAATGAAGACCCCTCGTGCGATGCTCGGTATCTGCGGTGTATTGAACAAAGGCATGTCCGGTGTTACACTAGTCTACATTCTTCTTGGCTTCCTCGGCTACCTGCGGTTCGGAGAAGAAGTCCAAGATTCCATCACTCTTAGTTTAGGGGACGCGATCCCTGCTCAAATAGTAAAACTGTCTATAGCCATTGCAGTATACTGCACTTTTGGCCTGCAGTTCTTCGTGTGCATTGATATTATGTGGAACGGTATTAAGGATAAATTCACGAAACGACCGGAGCTCGCCGATTACATCATGAGAACGATTATGGTCACGGTGTGCGTGTCGATTGCCGCGGCCGTTCCCCAGATTAGTCCTTTGATGGGAGTCATTGGCGCATTCTGTTTCTCCATCCTCGGTCTTATCGCACCAGCAATGATTGAAGTCATTACGTATTGGGAACCGGGAGTCGGCCCAGGAAAGTATTTGATCTGGAAAAACATTCTAGTGGTTATTTTTGGTACATTCTCCTTAGTTTTTGGTACTAAAGACGCCATACTTGCAATTTTAAAAGACTTCTAA
Protein
MVNEPNGAIPAPQELETFISPDEKNKEKIVSKYNMTKDAESGDFDPFTERKLDTPTSNMDTLTHLLKASLGTGILAMPKAFKAAGLINGILFTVIVAVVCTHCSYILVKCAHVLYKKTKKTQMSFAEVGEAALDNGPQPLRKWANAFRIFIVISLFITYFGTCSVYTVIIARNILQIIQFYCGDIINERFLIIILLVPLILMAWIRNLKYLAPVSMIANLFMAVGLGITLYYLVGTGQLQYDKIKDMLFKHPSEWPEFFSLTIFAMEAIGVVMPLENSMKTPRAMLGICGVLNKGMSGVTLVYILLGFLGYLRFGEEVQDSITLSLGDAIPAQIVKLSIAIAVYCTFGLQFFVCIDIMWNGIKDKFTKRPELADYIMRTIMVTVCVSIAAAVPQISPLMGVIGAFCFSILGLIAPAMIEVITYWEPGVGPGKYLIWKNILVVIFGTFSLVFGTKDAILAILKDF
Summary
Uniprot
H9ITY4
A0A2H1W139
A0A1E1W1L5
A0A194QLB1
A0A194PG23
S4PVP2
+ More
A0A3G1T1N4 A0A212EGM1 A0A0L7LDU2 A0A182I3Q1 A0A182V5K5 A0A182TIK4 A0A182KRU0 A0A182X6E6 Q7QJS2 A0A182NHK9 A0A182XZ97 A0A182QN38 A0A182WIS1 A0A182LSS4 T1DUB2 A0A2M4CTK1 A0A2M4A162 A7UR81 A0A2M3ZFK7 A0A2M4BMD6 A0A2M4BMF4 A0A154PAV5 A0A182JNY1 B0W3P2 A0A088A551 A0A310S4I8 A0A0M9A5G6 V9IEX3 U5EH92 A0A2A3E6U0 A0A0K8TMV6 A0A336LRJ5 A0A182P3J2 A0A1Q3FHL4 A0A182ILE4 A0A336KQA5 A0A0P6J1D0 A0A023ESM7 A0A0L7R832 A0A182T8X6 A0A336M521 A0A026WVX6 A0A1L8DEQ1 A0A182RFP0 A0A336K4X9 A0A0J7P0D7 E9IEM6 W5JB02 A0A2J7RDD7 A0A2J7RDD8 A0A182FI98 B4IY78 A0A2M4CTR5 A0A1J1HFU1 A0A195FPZ4 B4KVS3 W8B263 T1D3Z2 A0A158NEV2 A0A195BWB0 T1E213 A0A2A3E5B0 A0A0K8U669 A0A0L0C5S0 A0A1I8NWA9 A0A1Q3FHP5 B4LI66 A0A0A1X3D3 A0A195CAL6 A0A151WSS9 F4WJC9 E2A7X2 E2B643 A0A232F0F3 T1PDN1 A0A0M5JBI8 A0A151J0B4 A0A3B0JT44 A0A0Q9XCA9 A0A0Q9WIV5 Q29EE4 A0A1B6DLW5 A0A0K8VMX2 A0A0A1X926 B4N4M8 A0A084VFC4 A0A0R1DY79 A0A1A9UG14 A0A1A9ZKH4 A0A1B0FBH7 A0A3B0JMZ8 A0A0R3P676 B3NCH9 A0A1A9Y056
A0A3G1T1N4 A0A212EGM1 A0A0L7LDU2 A0A182I3Q1 A0A182V5K5 A0A182TIK4 A0A182KRU0 A0A182X6E6 Q7QJS2 A0A182NHK9 A0A182XZ97 A0A182QN38 A0A182WIS1 A0A182LSS4 T1DUB2 A0A2M4CTK1 A0A2M4A162 A7UR81 A0A2M3ZFK7 A0A2M4BMD6 A0A2M4BMF4 A0A154PAV5 A0A182JNY1 B0W3P2 A0A088A551 A0A310S4I8 A0A0M9A5G6 V9IEX3 U5EH92 A0A2A3E6U0 A0A0K8TMV6 A0A336LRJ5 A0A182P3J2 A0A1Q3FHL4 A0A182ILE4 A0A336KQA5 A0A0P6J1D0 A0A023ESM7 A0A0L7R832 A0A182T8X6 A0A336M521 A0A026WVX6 A0A1L8DEQ1 A0A182RFP0 A0A336K4X9 A0A0J7P0D7 E9IEM6 W5JB02 A0A2J7RDD7 A0A2J7RDD8 A0A182FI98 B4IY78 A0A2M4CTR5 A0A1J1HFU1 A0A195FPZ4 B4KVS3 W8B263 T1D3Z2 A0A158NEV2 A0A195BWB0 T1E213 A0A2A3E5B0 A0A0K8U669 A0A0L0C5S0 A0A1I8NWA9 A0A1Q3FHP5 B4LI66 A0A0A1X3D3 A0A195CAL6 A0A151WSS9 F4WJC9 E2A7X2 E2B643 A0A232F0F3 T1PDN1 A0A0M5JBI8 A0A151J0B4 A0A3B0JT44 A0A0Q9XCA9 A0A0Q9WIV5 Q29EE4 A0A1B6DLW5 A0A0K8VMX2 A0A0A1X926 B4N4M8 A0A084VFC4 A0A0R1DY79 A0A1A9UG14 A0A1A9ZKH4 A0A1B0FBH7 A0A3B0JMZ8 A0A0R3P676 B3NCH9 A0A1A9Y056
Pubmed
EMBL
BABH01007164
ODYU01005657
SOQ46733.1
GDQN01010188
GDQN01007066
JAT80866.1
+ More
JAT83988.1 KQ461196 KPJ06322.1 KQ459605 KPI91978.1 GAIX01008193 JAA84367.1 MG992469 AXY94907.1 AGBW02015069 OWR40620.1 JTDY01001553 KOB73554.1 APCN01000217 AAAB01008807 EAA04057.4 EDO64522.1 EDO64523.1 EDO64524.1 AXCN02000593 AXCM01000671 GAMD01000309 JAB01282.1 GGFL01004475 MBW68653.1 GGFK01001235 MBW34556.1 EDO64525.1 GGFM01006459 MBW27210.1 GGFJ01005051 MBW54192.1 GGFJ01005071 MBW54212.1 KQ434846 KZC08358.1 DS231833 EDS32189.1 KQ777311 OAD52142.1 KQ435729 KOX77627.1 JR042358 AEY59605.1 GANO01003126 JAB56745.1 KZ288364 PBC26879.1 GDAI01001924 JAI15679.1 UFQS01000120 UFQT01000120 SSX00035.1 SSX20415.1 GFDL01008026 JAV27019.1 AXCP01008206 UFQS01000356 UFQT01000356 SSX03120.1 SSX23486.1 GDUN01000062 JAN95857.1 GAPW01001325 JAC12273.1 KQ414633 KOC67042.1 SSX03121.1 SSX23487.1 KK107079 QOIP01000010 EZA60157.1 RLU17829.1 GFDF01009166 JAV04918.1 SSX00034.1 SSX20414.1 LBMM01000545 KMQ98090.1 GL762647 EFZ20983.1 ADMH02001751 ETN61181.1 NEVH01005291 PNF38842.1 PNF38840.1 CH916366 EDV96528.1 GGFL01004532 MBW68710.1 CVRI01000002 CRK86864.1 KQ981335 KYN42665.1 CH933809 EDW18447.1 GAMC01019199 JAB87356.1 GALA01001158 JAA93694.1 ADTU01013180 KQ976401 KYM92570.1 GALA01001159 JAA93693.1 PBC26878.1 GDHF01030479 GDHF01004180 JAI21835.1 JAI48134.1 JRES01000850 KNC27738.1 GFDL01008012 JAV27033.1 CH940647 EDW69633.1 GBXI01013416 GBXI01008650 JAD00876.1 JAD05642.1 KQ978023 KYM97914.1 KQ982769 KYQ50851.1 GL888182 EGI65707.1 GL437442 EFN70451.1 GL445914 EFN88832.1 NNAY01001356 OXU24256.1 KA646892 AFP61521.1 CP012525 ALC44494.1 KQ980636 KYN14945.1 OUUW01000002 SPP76859.1 KRG06192.1 KRF84490.1 CH379070 EAL30117.1 GEDC01010629 JAS26669.1 GDHF01012095 JAI40219.1 GBXI01006896 JAD07396.1 CH964095 EDW79102.2 ATLV01012374 KE524785 KFB36668.1 CM000159 KRK00867.1 CCAG010019568 SPP76860.1 KRT08657.1 CH954178 EDV51209.2
JAT83988.1 KQ461196 KPJ06322.1 KQ459605 KPI91978.1 GAIX01008193 JAA84367.1 MG992469 AXY94907.1 AGBW02015069 OWR40620.1 JTDY01001553 KOB73554.1 APCN01000217 AAAB01008807 EAA04057.4 EDO64522.1 EDO64523.1 EDO64524.1 AXCN02000593 AXCM01000671 GAMD01000309 JAB01282.1 GGFL01004475 MBW68653.1 GGFK01001235 MBW34556.1 EDO64525.1 GGFM01006459 MBW27210.1 GGFJ01005051 MBW54192.1 GGFJ01005071 MBW54212.1 KQ434846 KZC08358.1 DS231833 EDS32189.1 KQ777311 OAD52142.1 KQ435729 KOX77627.1 JR042358 AEY59605.1 GANO01003126 JAB56745.1 KZ288364 PBC26879.1 GDAI01001924 JAI15679.1 UFQS01000120 UFQT01000120 SSX00035.1 SSX20415.1 GFDL01008026 JAV27019.1 AXCP01008206 UFQS01000356 UFQT01000356 SSX03120.1 SSX23486.1 GDUN01000062 JAN95857.1 GAPW01001325 JAC12273.1 KQ414633 KOC67042.1 SSX03121.1 SSX23487.1 KK107079 QOIP01000010 EZA60157.1 RLU17829.1 GFDF01009166 JAV04918.1 SSX00034.1 SSX20414.1 LBMM01000545 KMQ98090.1 GL762647 EFZ20983.1 ADMH02001751 ETN61181.1 NEVH01005291 PNF38842.1 PNF38840.1 CH916366 EDV96528.1 GGFL01004532 MBW68710.1 CVRI01000002 CRK86864.1 KQ981335 KYN42665.1 CH933809 EDW18447.1 GAMC01019199 JAB87356.1 GALA01001158 JAA93694.1 ADTU01013180 KQ976401 KYM92570.1 GALA01001159 JAA93693.1 PBC26878.1 GDHF01030479 GDHF01004180 JAI21835.1 JAI48134.1 JRES01000850 KNC27738.1 GFDL01008012 JAV27033.1 CH940647 EDW69633.1 GBXI01013416 GBXI01008650 JAD00876.1 JAD05642.1 KQ978023 KYM97914.1 KQ982769 KYQ50851.1 GL888182 EGI65707.1 GL437442 EFN70451.1 GL445914 EFN88832.1 NNAY01001356 OXU24256.1 KA646892 AFP61521.1 CP012525 ALC44494.1 KQ980636 KYN14945.1 OUUW01000002 SPP76859.1 KRG06192.1 KRF84490.1 CH379070 EAL30117.1 GEDC01010629 JAS26669.1 GDHF01012095 JAI40219.1 GBXI01006896 JAD07396.1 CH964095 EDW79102.2 ATLV01012374 KE524785 KFB36668.1 CM000159 KRK00867.1 CCAG010019568 SPP76860.1 KRT08657.1 CH954178 EDV51209.2
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000037510
UP000075840
+ More
UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075884 UP000076408 UP000075886 UP000075920 UP000075883 UP000076502 UP000075881 UP000002320 UP000005203 UP000053105 UP000242457 UP000075885 UP000075880 UP000053825 UP000075901 UP000053097 UP000279307 UP000075900 UP000036403 UP000000673 UP000235965 UP000069272 UP000001070 UP000183832 UP000078541 UP000009192 UP000005205 UP000078540 UP000037069 UP000095300 UP000008792 UP000078542 UP000075809 UP000007755 UP000000311 UP000008237 UP000215335 UP000095301 UP000092553 UP000078492 UP000268350 UP000001819 UP000007798 UP000030765 UP000002282 UP000078200 UP000092445 UP000092444 UP000008711 UP000092443
UP000075903 UP000075902 UP000075882 UP000076407 UP000007062 UP000075884 UP000076408 UP000075886 UP000075920 UP000075883 UP000076502 UP000075881 UP000002320 UP000005203 UP000053105 UP000242457 UP000075885 UP000075880 UP000053825 UP000075901 UP000053097 UP000279307 UP000075900 UP000036403 UP000000673 UP000235965 UP000069272 UP000001070 UP000183832 UP000078541 UP000009192 UP000005205 UP000078540 UP000037069 UP000095300 UP000008792 UP000078542 UP000075809 UP000007755 UP000000311 UP000008237 UP000215335 UP000095301 UP000092553 UP000078492 UP000268350 UP000001819 UP000007798 UP000030765 UP000002282 UP000078200 UP000092445 UP000092444 UP000008711 UP000092443
PRIDE
Pfam
PF01490 Aa_trans
Interpro
IPR013057
AA_transpt_TM
ProteinModelPortal
H9ITY4
A0A2H1W139
A0A1E1W1L5
A0A194QLB1
A0A194PG23
S4PVP2
+ More
A0A3G1T1N4 A0A212EGM1 A0A0L7LDU2 A0A182I3Q1 A0A182V5K5 A0A182TIK4 A0A182KRU0 A0A182X6E6 Q7QJS2 A0A182NHK9 A0A182XZ97 A0A182QN38 A0A182WIS1 A0A182LSS4 T1DUB2 A0A2M4CTK1 A0A2M4A162 A7UR81 A0A2M3ZFK7 A0A2M4BMD6 A0A2M4BMF4 A0A154PAV5 A0A182JNY1 B0W3P2 A0A088A551 A0A310S4I8 A0A0M9A5G6 V9IEX3 U5EH92 A0A2A3E6U0 A0A0K8TMV6 A0A336LRJ5 A0A182P3J2 A0A1Q3FHL4 A0A182ILE4 A0A336KQA5 A0A0P6J1D0 A0A023ESM7 A0A0L7R832 A0A182T8X6 A0A336M521 A0A026WVX6 A0A1L8DEQ1 A0A182RFP0 A0A336K4X9 A0A0J7P0D7 E9IEM6 W5JB02 A0A2J7RDD7 A0A2J7RDD8 A0A182FI98 B4IY78 A0A2M4CTR5 A0A1J1HFU1 A0A195FPZ4 B4KVS3 W8B263 T1D3Z2 A0A158NEV2 A0A195BWB0 T1E213 A0A2A3E5B0 A0A0K8U669 A0A0L0C5S0 A0A1I8NWA9 A0A1Q3FHP5 B4LI66 A0A0A1X3D3 A0A195CAL6 A0A151WSS9 F4WJC9 E2A7X2 E2B643 A0A232F0F3 T1PDN1 A0A0M5JBI8 A0A151J0B4 A0A3B0JT44 A0A0Q9XCA9 A0A0Q9WIV5 Q29EE4 A0A1B6DLW5 A0A0K8VMX2 A0A0A1X926 B4N4M8 A0A084VFC4 A0A0R1DY79 A0A1A9UG14 A0A1A9ZKH4 A0A1B0FBH7 A0A3B0JMZ8 A0A0R3P676 B3NCH9 A0A1A9Y056
A0A3G1T1N4 A0A212EGM1 A0A0L7LDU2 A0A182I3Q1 A0A182V5K5 A0A182TIK4 A0A182KRU0 A0A182X6E6 Q7QJS2 A0A182NHK9 A0A182XZ97 A0A182QN38 A0A182WIS1 A0A182LSS4 T1DUB2 A0A2M4CTK1 A0A2M4A162 A7UR81 A0A2M3ZFK7 A0A2M4BMD6 A0A2M4BMF4 A0A154PAV5 A0A182JNY1 B0W3P2 A0A088A551 A0A310S4I8 A0A0M9A5G6 V9IEX3 U5EH92 A0A2A3E6U0 A0A0K8TMV6 A0A336LRJ5 A0A182P3J2 A0A1Q3FHL4 A0A182ILE4 A0A336KQA5 A0A0P6J1D0 A0A023ESM7 A0A0L7R832 A0A182T8X6 A0A336M521 A0A026WVX6 A0A1L8DEQ1 A0A182RFP0 A0A336K4X9 A0A0J7P0D7 E9IEM6 W5JB02 A0A2J7RDD7 A0A2J7RDD8 A0A182FI98 B4IY78 A0A2M4CTR5 A0A1J1HFU1 A0A195FPZ4 B4KVS3 W8B263 T1D3Z2 A0A158NEV2 A0A195BWB0 T1E213 A0A2A3E5B0 A0A0K8U669 A0A0L0C5S0 A0A1I8NWA9 A0A1Q3FHP5 B4LI66 A0A0A1X3D3 A0A195CAL6 A0A151WSS9 F4WJC9 E2A7X2 E2B643 A0A232F0F3 T1PDN1 A0A0M5JBI8 A0A151J0B4 A0A3B0JT44 A0A0Q9XCA9 A0A0Q9WIV5 Q29EE4 A0A1B6DLW5 A0A0K8VMX2 A0A0A1X926 B4N4M8 A0A084VFC4 A0A0R1DY79 A0A1A9UG14 A0A1A9ZKH4 A0A1B0FBH7 A0A3B0JMZ8 A0A0R3P676 B3NCH9 A0A1A9Y056
Ontologies
GO
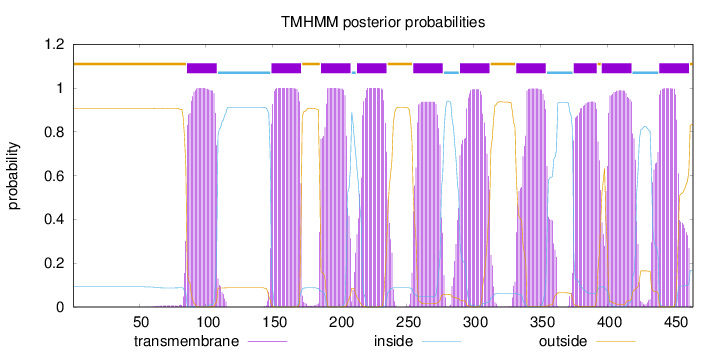
Topology
Length:
464
Number of predicted TMHs:
10
Exp number of AAs in TMHs:
214.2835
Exp number, first 60 AAs:
0.00286
Total prob of N-in:
0.09201
outside
1 - 85
TMhelix
86 - 108
inside
109 - 148
TMhelix
149 - 171
outside
172 - 185
TMhelix
186 - 208
inside
209 - 212
TMhelix
213 - 235
outside
236 - 254
TMhelix
255 - 277
inside
278 - 289
TMhelix
290 - 312
outside
313 - 331
TMhelix
332 - 354
inside
355 - 374
TMhelix
375 - 392
outside
393 - 395
TMhelix
396 - 418
inside
419 - 438
TMhelix
439 - 461
outside
462 - 464
Population Genetic Test Statistics
Pi
20.278061
Theta
19.161021
Tajima's D
0.17086
CLR
0.014802
CSRT
0.426278686065697
Interpretation
Uncertain
