Gene
KWMTBOMO00366 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000516
Annotation
PREDICTED:_insulin-like_growth_factor_2_mRNA-binding_protein_1_isoform_X1_[Bombyx_mori]
Location in the cell
Nuclear Reliability : 3.589
Sequence
CDS
ATGTCTAATCCAATGGAGCAACAGTTCGAGCTTAATTTATCACAAGAGGACCATGATCAAATATTTGAACAAGACGATCTCCACGATCAAAGATCCTCAAAGGTACTCATCAGTGGGCTACCTCTGCATATCCGCTTCGATAACATCGAGCCACTGCTCTCACAGTATGGCAACGTGCAGCATTGCGACAAAGCCAACACCCGCGATCCCAACACCCAAGCGGTGTTCATCACCTTCGAGAGTCCAGAGCAGGCGCAGCAAGCCATAAACGGGTTGAATGGATGTGAATTGGAGGGTTGTCGTATAAAAGTGGAGGCAGCAGAGCAGAATGCTCGTGGAGGTCGTCGCGGTCGCCCTGGTGGCGGCCGTGGTGGAGGCGGCGCTGCTGGCGGTGGCTCGCGCCCCACCGACTTCCCACTGCGTCTGCTGGTGCAGAGCGACATGGTCGGCGCTATCATCGGTCGCCAGGGCAGTACCATTCGACTCATCACCCAGCAGAGTCGTGCTCGCGTTGACGTACACAGGAAGGATAATGTCGGCTCCCTGGAAAAAGCAATTACCATCTATGGGAATCCCGAGAATTGCACCAATGCCTGCAAGAGAATCCTTGAAGTAATGCAGCAGGAGGCCAATAATACTAATAAAGGCGAAATATGCCTGAAGATTTTGGCCCATAATAATCTCATTGGACGCATCATCGGCAAAGGCGGCAACACCATCAAGAGAATTATGCAAGAGACCGATACAAAGATCACTGTCTCCTCTATAAATGATATCAATAGCTTCAATTTGGAGAGAATCATAACCGTTAAGGGCTCCATCGAGAACATGGCCAAGGCGGAATCGCAAATTTCAGCCAAATTGCGTCAAAGCTACGAAAATGATTTGCAGGTGCTGGCGCCGCAAAGCATTATGTTCCCTGGGTTGCACCCAATGGCTATGATGTCGACTGGACGTGGTTTCTGTGGAGCTCCGCCACCGTTCCAGCCGCCGATGTATGCGCCGCTGGCCGGCCCTGGGGCACAGCAGGGTGCCGGTGACTCGCAGGAAACAACCTATCTGTACATCCCAAACAACGCTGTGGGCGCGATTATCGGTACAAAGGGCTCGCACATTCGCAACATTATTAGATTCAGCAATGCTTCTGTTAAGATTGCGCCTTTGGAACAAGAAAAGCAACAGGAACCGCAGAACCCACAGCAAGAGCGCAAGGTTACCATTGTCGGTAGTCCTGAGGCGCAATGGAAGGCGCAGTATCTTATCTTTGAAAAGATGCGGGAAGAAGGTTTTATGTCTGGATCGGATGACGTGCGGCTGATTGTGGAAATTGTCGTGGCCTCGTCGCAAGTGGGTCGGATCATCGGCAAAGGCGGACAGAACGTGAGGGAGCTTCAGCGCGTGACCGGCTCGCTGATCAAGCTACCCGAGCAGCAGCAGCAGCCCGGCTCGCAGGGACAAGAGCACGAGACCACCGTTCACATTGTGGGCCCCTTTTATAGCGTCCAGTCTGCGCAGCGACGCATCCGTGCCATGGTGGCGCAGGCGAGCGCGCCGGGACGCCGCCGCGCCGCGCAGCCCCCGCCCGTGCAGCAGTAG
Protein
MSNPMEQQFELNLSQEDHDQIFEQDDLHDQRSSKVLISGLPLHIRFDNIEPLLSQYGNVQHCDKANTRDPNTQAVFITFESPEQAQQAINGLNGCELEGCRIKVEAAEQNARGGRRGRPGGGRGGGGAAGGGSRPTDFPLRLLVQSDMVGAIIGRQGSTIRLITQQSRARVDVHRKDNVGSLEKAITIYGNPENCTNACKRILEVMQQEANNTNKGEICLKILAHNNLIGRIIGKGGNTIKRIMQETDTKITVSSINDINSFNLERIITVKGSIENMAKAESQISAKLRQSYENDLQVLAPQSIMFPGLHPMAMMSTGRGFCGAPPPFQPPMYAPLAGPGAQQGAGDSQETTYLYIPNNAVGAIIGTKGSHIRNIIRFSNASVKIAPLEQEKQQEPQNPQQERKVTIVGSPEAQWKAQYLIFEKMREEGFMSGSDDVRLIVEIVVASSQVGRIIGKGGQNVRELQRVTGSLIKLPEQQQQPGSQGQEHETTVHIVGPFYSVQSAQRRIRAMVAQASAPGRRRAAQPPPVQQ
Summary
Uniprot
A0A2W1BI28
A0A212EY18
A0A194QUW5
A0A194PTG8
A0A0L7L7I0
H9ITD6
+ More
A0A139WIL4 A0A1Y1N798 A0A182GJ16 U5EWF3 A0A182HC23 A0A139WI47 A0A0P6IX28 A0A1Q3G072 A0A1Q3G096 A0A1Q3G077 A0A1Q3G050 Q174K3 T1D3W6 A0A1Y1N760 A0A1J1J6D3 B4M2G9 A0A182LSY7 A0A1W4UNZ0 A0A1I8P5R7 A0A1I8MKW2 A0A182RAI2 A0A182KT96 A0A182P1F5 A0A182X068 A0A0Q5T3E7 A0A182HPL0 A0A182N3R4 A0A182YC90 A0A182JBB4 A0A182WD34 A0A1B6FVV0 A0A084WIY1 A0A1A9X576 A0A182QRV3 B0WBU9 R4WDQ2 A0A0Q5T5Z7 A0A1W4V184 A0A158NU30 F4WK49 A0A1I8MKW7 A0A151I4M8 A0A0R1EC75 A0A1I8P5M4 A0A1I8P5S9 A0A195DJL8 A0A151X023 A0A1B0FL37 U4U949 A0A0K8W218 M9NF14 A0A034VZ55 A0A026X2G9 A0A3L8DWB3 E9IRT0 A0A0M3QZ36 A0A182UJW1 K7J7K6 A0A3F2Z096 A0A0R1EJ14 A0A0Q5T351 W8BY82 N6THE1 E2C260 A0A0C9RYT5 A0A1B6FSQ9 A0A182S8Z5 A0A1A9XBV9 A0A0K8UZ75 A0A1B0B896 A0A034VWH9 E1ZZ14 A0A0A1WHJ7 A0A034W019 Q0KHU2 A0A1A9VRJ7 A0A034W0J6 A0A067RX66 A0A069DVP4 T1H969 A0A023FAG8 A0A224XE40 A0A2R7VU76 A0A2R7VTU5 A0A1A9ZGM2 A0A2P8YLL4 Q29GU3 A0A182KBX9 A0A195C988 A0A146LUH0 A0A1B6GML6 A0A2M4CQS7 E0VXF6 A0A154PTH1
A0A139WIL4 A0A1Y1N798 A0A182GJ16 U5EWF3 A0A182HC23 A0A139WI47 A0A0P6IX28 A0A1Q3G072 A0A1Q3G096 A0A1Q3G077 A0A1Q3G050 Q174K3 T1D3W6 A0A1Y1N760 A0A1J1J6D3 B4M2G9 A0A182LSY7 A0A1W4UNZ0 A0A1I8P5R7 A0A1I8MKW2 A0A182RAI2 A0A182KT96 A0A182P1F5 A0A182X068 A0A0Q5T3E7 A0A182HPL0 A0A182N3R4 A0A182YC90 A0A182JBB4 A0A182WD34 A0A1B6FVV0 A0A084WIY1 A0A1A9X576 A0A182QRV3 B0WBU9 R4WDQ2 A0A0Q5T5Z7 A0A1W4V184 A0A158NU30 F4WK49 A0A1I8MKW7 A0A151I4M8 A0A0R1EC75 A0A1I8P5M4 A0A1I8P5S9 A0A195DJL8 A0A151X023 A0A1B0FL37 U4U949 A0A0K8W218 M9NF14 A0A034VZ55 A0A026X2G9 A0A3L8DWB3 E9IRT0 A0A0M3QZ36 A0A182UJW1 K7J7K6 A0A3F2Z096 A0A0R1EJ14 A0A0Q5T351 W8BY82 N6THE1 E2C260 A0A0C9RYT5 A0A1B6FSQ9 A0A182S8Z5 A0A1A9XBV9 A0A0K8UZ75 A0A1B0B896 A0A034VWH9 E1ZZ14 A0A0A1WHJ7 A0A034W019 Q0KHU2 A0A1A9VRJ7 A0A034W0J6 A0A067RX66 A0A069DVP4 T1H969 A0A023FAG8 A0A224XE40 A0A2R7VU76 A0A2R7VTU5 A0A1A9ZGM2 A0A2P8YLL4 Q29GU3 A0A182KBX9 A0A195C988 A0A146LUH0 A0A1B6GML6 A0A2M4CQS7 E0VXF6 A0A154PTH1
Pubmed
28756777
22118469
26354079
26227816
19121390
18362917
+ More
19820115 28004739 26483478 26999592 17510324 24330624 17994087 25315136 20966253 25244985 24438588 23691247 21347285 21719571 17550304 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 24508170 30249741 21282665 20075255 24495485 20798317 25830018 24845553 26334808 25474469 29403074 15632085 26823975 20566863
19820115 28004739 26483478 26999592 17510324 24330624 17994087 25315136 20966253 25244985 24438588 23691247 21347285 21719571 17550304 23537049 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25348373 24508170 30249741 21282665 20075255 24495485 20798317 25830018 24845553 26334808 25474469 29403074 15632085 26823975 20566863
EMBL
KZ150032
PZC74702.1
AGBW02011631
OWR46380.1
KQ461108
KPJ09323.1
+ More
KQ459594 KPI96054.1 JTDY01002518 KOB71246.1 BABH01007153 KQ971342 KYB27607.1 GEZM01010855 GEZM01010851 JAV93763.1 JXUM01067178 KQ562437 KXJ75915.1 GANO01002972 JAB56899.1 JXUM01033215 JXUM01033216 JXUM01033217 JXUM01033218 JXUM01033219 JXUM01033220 JXUM01033221 JXUM01033222 KQ560981 KXJ80149.1 KYB27606.1 GDUN01000515 JAN95404.1 GFDL01001852 JAV33193.1 GFDL01001849 JAV33196.1 GFDL01001851 JAV33194.1 GFDL01001850 JAV33195.1 CH477409 EAT41502.1 GALA01001208 JAA93644.1 GEZM01010854 JAV93764.1 CVRI01000074 CRL07971.1 CH940651 EDW65873.1 AXCM01003862 CH954180 KQS29837.1 APCN01003264 APCN01003265 GECZ01015442 JAS54327.1 ATLV01023952 KE525347 KFB50175.1 AXCN02001101 DS231882 EDS42879.1 AK417739 BAN20954.1 KQS29840.1 ADTU01026256 ADTU01026257 ADTU01026258 ADTU01026259 ADTU01026260 ADTU01026261 ADTU01026262 ADTU01026263 GL888193 EGI65421.1 KQ976445 KYM86030.1 CM000162 KRK07051.1 KQ980794 KYN13078.1 KQ982638 KYQ53275.1 CCAG010022093 CCAG010022094 CCAG010022095 KB631923 ERL87146.1 GDHF01028931 GDHF01010265 GDHF01007193 JAI23383.1 JAI42049.1 JAI45121.1 AE014298 AFH07330.1 AHN59561.1 GAKP01011275 GAKP01011273 JAC47679.1 KK107020 EZA62482.1 QOIP01000004 RLU24068.1 GL765283 EFZ16735.1 CP012528 ALC48743.1 AAZX01002184 AAZX01008433 KRK07050.1 KQS29836.1 GAMC01004997 JAC01559.1 APGK01037933 KB740949 ENN77188.1 GL452067 EFN78011.1 GBYB01014490 JAG84257.1 GECZ01016539 JAS53230.1 GDHF01020418 JAI31896.1 JXJN01009884 GAKP01011276 JAC47676.1 GL435250 EFN73581.1 GBXI01015763 JAC98528.1 GAKP01011280 JAC47672.1 ABI30974.2 GAKP01011277 JAC47675.1 KK852409 KDR24489.1 GBGD01001018 JAC87871.1 ACPB03007560 GBBI01000231 JAC18481.1 GFTR01007162 JAW09264.1 KK854084 PTY10919.1 PTY10917.1 PYGN01000509 PSN45142.1 CH379064 EAL32016.3 KQ978143 KYM96736.1 GDHC01008292 JAQ10337.1 GECZ01006077 JAS63692.1 GGFL01003070 MBW67248.1 DS235830 EEB18062.1 KQ435095 KZC14648.1
KQ459594 KPI96054.1 JTDY01002518 KOB71246.1 BABH01007153 KQ971342 KYB27607.1 GEZM01010855 GEZM01010851 JAV93763.1 JXUM01067178 KQ562437 KXJ75915.1 GANO01002972 JAB56899.1 JXUM01033215 JXUM01033216 JXUM01033217 JXUM01033218 JXUM01033219 JXUM01033220 JXUM01033221 JXUM01033222 KQ560981 KXJ80149.1 KYB27606.1 GDUN01000515 JAN95404.1 GFDL01001852 JAV33193.1 GFDL01001849 JAV33196.1 GFDL01001851 JAV33194.1 GFDL01001850 JAV33195.1 CH477409 EAT41502.1 GALA01001208 JAA93644.1 GEZM01010854 JAV93764.1 CVRI01000074 CRL07971.1 CH940651 EDW65873.1 AXCM01003862 CH954180 KQS29837.1 APCN01003264 APCN01003265 GECZ01015442 JAS54327.1 ATLV01023952 KE525347 KFB50175.1 AXCN02001101 DS231882 EDS42879.1 AK417739 BAN20954.1 KQS29840.1 ADTU01026256 ADTU01026257 ADTU01026258 ADTU01026259 ADTU01026260 ADTU01026261 ADTU01026262 ADTU01026263 GL888193 EGI65421.1 KQ976445 KYM86030.1 CM000162 KRK07051.1 KQ980794 KYN13078.1 KQ982638 KYQ53275.1 CCAG010022093 CCAG010022094 CCAG010022095 KB631923 ERL87146.1 GDHF01028931 GDHF01010265 GDHF01007193 JAI23383.1 JAI42049.1 JAI45121.1 AE014298 AFH07330.1 AHN59561.1 GAKP01011275 GAKP01011273 JAC47679.1 KK107020 EZA62482.1 QOIP01000004 RLU24068.1 GL765283 EFZ16735.1 CP012528 ALC48743.1 AAZX01002184 AAZX01008433 KRK07050.1 KQS29836.1 GAMC01004997 JAC01559.1 APGK01037933 KB740949 ENN77188.1 GL452067 EFN78011.1 GBYB01014490 JAG84257.1 GECZ01016539 JAS53230.1 GDHF01020418 JAI31896.1 JXJN01009884 GAKP01011276 JAC47676.1 GL435250 EFN73581.1 GBXI01015763 JAC98528.1 GAKP01011280 JAC47672.1 ABI30974.2 GAKP01011277 JAC47675.1 KK852409 KDR24489.1 GBGD01001018 JAC87871.1 ACPB03007560 GBBI01000231 JAC18481.1 GFTR01007162 JAW09264.1 KK854084 PTY10919.1 PTY10917.1 PYGN01000509 PSN45142.1 CH379064 EAL32016.3 KQ978143 KYM96736.1 GDHC01008292 JAQ10337.1 GECZ01006077 JAS63692.1 GGFL01003070 MBW67248.1 DS235830 EEB18062.1 KQ435095 KZC14648.1
Proteomes
UP000007151
UP000053240
UP000053268
UP000037510
UP000005204
UP000007266
+ More
UP000069940 UP000249989 UP000008820 UP000183832 UP000008792 UP000075883 UP000192221 UP000095300 UP000095301 UP000075900 UP000075882 UP000075885 UP000076407 UP000008711 UP000075840 UP000075884 UP000076408 UP000075880 UP000075920 UP000030765 UP000091820 UP000075886 UP000002320 UP000005205 UP000007755 UP000078540 UP000002282 UP000078492 UP000075809 UP000092444 UP000030742 UP000000803 UP000053097 UP000279307 UP000092553 UP000075902 UP000002358 UP000075903 UP000019118 UP000008237 UP000075901 UP000092443 UP000092460 UP000000311 UP000078200 UP000027135 UP000015103 UP000092445 UP000245037 UP000001819 UP000075881 UP000078542 UP000009046 UP000076502
UP000069940 UP000249989 UP000008820 UP000183832 UP000008792 UP000075883 UP000192221 UP000095300 UP000095301 UP000075900 UP000075882 UP000075885 UP000076407 UP000008711 UP000075840 UP000075884 UP000076408 UP000075880 UP000075920 UP000030765 UP000091820 UP000075886 UP000002320 UP000005205 UP000007755 UP000078540 UP000002282 UP000078492 UP000075809 UP000092444 UP000030742 UP000000803 UP000053097 UP000279307 UP000092553 UP000075902 UP000002358 UP000075903 UP000019118 UP000008237 UP000075901 UP000092443 UP000092460 UP000000311 UP000078200 UP000027135 UP000015103 UP000092445 UP000245037 UP000001819 UP000075881 UP000078542 UP000009046 UP000076502
Interpro
Gene 3D
ProteinModelPortal
A0A2W1BI28
A0A212EY18
A0A194QUW5
A0A194PTG8
A0A0L7L7I0
H9ITD6
+ More
A0A139WIL4 A0A1Y1N798 A0A182GJ16 U5EWF3 A0A182HC23 A0A139WI47 A0A0P6IX28 A0A1Q3G072 A0A1Q3G096 A0A1Q3G077 A0A1Q3G050 Q174K3 T1D3W6 A0A1Y1N760 A0A1J1J6D3 B4M2G9 A0A182LSY7 A0A1W4UNZ0 A0A1I8P5R7 A0A1I8MKW2 A0A182RAI2 A0A182KT96 A0A182P1F5 A0A182X068 A0A0Q5T3E7 A0A182HPL0 A0A182N3R4 A0A182YC90 A0A182JBB4 A0A182WD34 A0A1B6FVV0 A0A084WIY1 A0A1A9X576 A0A182QRV3 B0WBU9 R4WDQ2 A0A0Q5T5Z7 A0A1W4V184 A0A158NU30 F4WK49 A0A1I8MKW7 A0A151I4M8 A0A0R1EC75 A0A1I8P5M4 A0A1I8P5S9 A0A195DJL8 A0A151X023 A0A1B0FL37 U4U949 A0A0K8W218 M9NF14 A0A034VZ55 A0A026X2G9 A0A3L8DWB3 E9IRT0 A0A0M3QZ36 A0A182UJW1 K7J7K6 A0A3F2Z096 A0A0R1EJ14 A0A0Q5T351 W8BY82 N6THE1 E2C260 A0A0C9RYT5 A0A1B6FSQ9 A0A182S8Z5 A0A1A9XBV9 A0A0K8UZ75 A0A1B0B896 A0A034VWH9 E1ZZ14 A0A0A1WHJ7 A0A034W019 Q0KHU2 A0A1A9VRJ7 A0A034W0J6 A0A067RX66 A0A069DVP4 T1H969 A0A023FAG8 A0A224XE40 A0A2R7VU76 A0A2R7VTU5 A0A1A9ZGM2 A0A2P8YLL4 Q29GU3 A0A182KBX9 A0A195C988 A0A146LUH0 A0A1B6GML6 A0A2M4CQS7 E0VXF6 A0A154PTH1
A0A139WIL4 A0A1Y1N798 A0A182GJ16 U5EWF3 A0A182HC23 A0A139WI47 A0A0P6IX28 A0A1Q3G072 A0A1Q3G096 A0A1Q3G077 A0A1Q3G050 Q174K3 T1D3W6 A0A1Y1N760 A0A1J1J6D3 B4M2G9 A0A182LSY7 A0A1W4UNZ0 A0A1I8P5R7 A0A1I8MKW2 A0A182RAI2 A0A182KT96 A0A182P1F5 A0A182X068 A0A0Q5T3E7 A0A182HPL0 A0A182N3R4 A0A182YC90 A0A182JBB4 A0A182WD34 A0A1B6FVV0 A0A084WIY1 A0A1A9X576 A0A182QRV3 B0WBU9 R4WDQ2 A0A0Q5T5Z7 A0A1W4V184 A0A158NU30 F4WK49 A0A1I8MKW7 A0A151I4M8 A0A0R1EC75 A0A1I8P5M4 A0A1I8P5S9 A0A195DJL8 A0A151X023 A0A1B0FL37 U4U949 A0A0K8W218 M9NF14 A0A034VZ55 A0A026X2G9 A0A3L8DWB3 E9IRT0 A0A0M3QZ36 A0A182UJW1 K7J7K6 A0A3F2Z096 A0A0R1EJ14 A0A0Q5T351 W8BY82 N6THE1 E2C260 A0A0C9RYT5 A0A1B6FSQ9 A0A182S8Z5 A0A1A9XBV9 A0A0K8UZ75 A0A1B0B896 A0A034VWH9 E1ZZ14 A0A0A1WHJ7 A0A034W019 Q0KHU2 A0A1A9VRJ7 A0A034W0J6 A0A067RX66 A0A069DVP4 T1H969 A0A023FAG8 A0A224XE40 A0A2R7VU76 A0A2R7VTU5 A0A1A9ZGM2 A0A2P8YLL4 Q29GU3 A0A182KBX9 A0A195C988 A0A146LUH0 A0A1B6GML6 A0A2M4CQS7 E0VXF6 A0A154PTH1
PDB
6QEY
E-value=9.0351e-50,
Score=498
Ontologies
GO
PANTHER
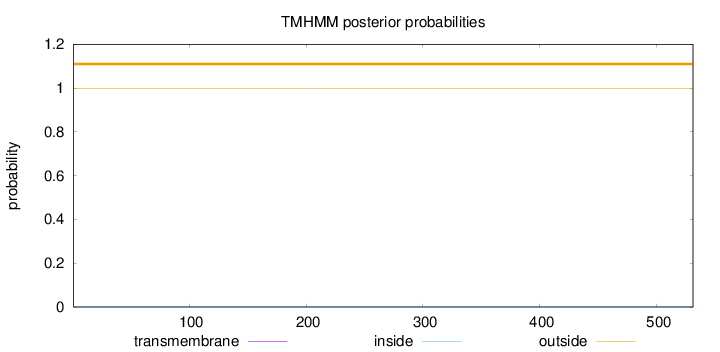
Topology
Length:
531
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0026
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00095
outside
1 - 531
Population Genetic Test Statistics
Pi
15.57594
Theta
18.712368
Tajima's D
-1.142349
CLR
0.045104
CSRT
0.109544522773861
Interpretation
Uncertain
