Gene
KWMTBOMO00364
Pre Gene Modal
BGIBMGA000711
Annotation
PREDICTED:_MAGUK_p55_subfamily_member_7_isoform_X2_[Amyelois_transitella]
Location in the cell
Cytoplasmic Reliability : 2.237
Sequence
CDS
ATGAACATGGATTCACCGTATGAAAATTGGGACCCAGCACTCACACGTTTGTTGACATCTTTGAAAGAGGTACAATCAGGTAGTGAAGATGTGGCTTTTCTTAGTGAACTGCTGCAGTCTAAGCAGTTGCATGCCTTGGTACATGTTCACAACAAAATTGTGGCTAAAGGCAAGGATGACAAATTTTATCCTCTACTCTCAAATGCTATGCAGGTTACACTTGAGGTTTTTGAAGTACTTGGTGATAAAACTAAAATTTCAAAAGAGTATGAAGAGTTACTAAGTTTATTGCAGAAGCCACATTTTCAGGCAATTTTGTGTACGCATGATGCAGTTGCTCAGAAAGACTATTACCCCCATCTACCTGATATTCCACCTGATACTGATGATGAAGAAGAAACTGTAAAGATTGTACAATTAGTCAAAAGTGATGAACCATTGGGCGCAACAATCAAGACTGACGAAGAAACCGGGAAGATAGTTATCGCTCGAGTGATGCACGGAGGTGCCGCTGATAGATCGGGACTCATCCACGCCGGCGACGAGGTCATCGAAGTTAACGGAATCAGTGTGGAAAACAAAACTCCGGCCGATGTCTTGTCTATATTACAAAACTCCGAAGGCACGATCACGTTTAAATTAGTACCGTCATTCGGTAAAGGCGGTACGCGGGAAAGCAAAGTCAGAGTCCGAGCGTTATTTGATTACAATTCGTCCGAAGATCCTTATATACCCTGTAAGGAAGCGGGACTCGATTTTAAGAAGGGTGACATCCTCCACATCGTTTCACAGGACGACGCCTACTGGTGGCAAGCGAGACGCGAAGGCGATAGAGTTATGAGGGCGGGGCTTATACCGTCCAGGGCTCTGCAAGAAGGTCGCATCATTCACGAGAGACAGACCGATCCCCAGACTATGGATGGTAAGCCCGCCCTTTGTAGTCCGTCTGCGTCCAGTTCTGACTGTGCCCCGAAAACGCCATGTTCACCGACGCCGACGGCCACAGCGCTCCTGCCCTGCAAGTCGATTCCGAAAGTCAAGAAGATCATATACGACATAACGGAGAACGACGATTTCGATAGAGAATTGATACCGACGTACGAGGAGGTCGCCCGGCTGTACCCGAGGCCGGGTCTGGTACGCCCGATCGTGTTGATCGGAGCGCCCGGCGTCGGCAGAAACGAACTGAGACGGAGGCTCGTCGCTACGGACCCGGAGAAGTACATCACTCCGATACCGTATACGTCGCGTCCTATAAAATCAAGCGAACAGAATGGTAAAGATTACGTGTTCGTAACTCGGGAAAAAATGGAACAGGATATAACCGATGGGAAATTCATAGAGCATGGTGAATACAAGGGCAATCTTTATGGGACCGCTGCCGAAAGCGTCGAGACTATTATTAATACTGGTCGTGTGTGTGTGTTGAGTCCGCATTGGCAGGCCCTAAAGATGCTTCGAACTCCGCGCCTGCGTCCCTACATCGTGTTCATTAAGCCTCCGTCCTTGGAGAGGATGGTCGAAACGCGGACTGCGGCAAACGCGCGTTCCACATTCGATAAGGAATCATCCAGAGCATTCACGGAAGAAGAGTTCACAGATATAATACGTTCGTCCAATCGTATTAATTTCTTATACGGATACATGTTCGATGAAGAGGTGGTGAATGAAGATCTGGCCAGTGCGGTGTCTCAACTACTCAAGGCCGCTTGGAGAGTTCAGTCTGAACCGCTGTGGGTACCTGCTTCATGGGTCCAGTAA
Protein
MNMDSPYENWDPALTRLLTSLKEVQSGSEDVAFLSELLQSKQLHALVHVHNKIVAKGKDDKFYPLLSNAMQVTLEVFEVLGDKTKISKEYEELLSLLQKPHFQAILCTHDAVAQKDYYPHLPDIPPDTDDEEETVKIVQLVKSDEPLGATIKTDEETGKIVIARVMHGGAADRSGLIHAGDEVIEVNGISVENKTPADVLSILQNSEGTITFKLVPSFGKGGTRESKVRVRALFDYNSSEDPYIPCKEAGLDFKKGDILHIVSQDDAYWWQARREGDRVMRAGLIPSRALQEGRIIHERQTDPQTMDGKPALCSPSASSSDCAPKTPCSPTPTATALLPCKSIPKVKKIIYDITENDDFDRELIPTYEEVARLYPRPGLVRPIVLIGAPGVGRNELRRRLVATDPEKYITPIPYTSRPIKSSEQNGKDYVFVTREKMEQDITDGKFIEHGEYKGNLYGTAAESVETIINTGRVCVLSPHWQALKMLRTPRLRPYIVFIKPPSLERMVETRTAANARSTFDKESSRAFTEEEFTDIIRSSNRINFLYGYMFDEEVVNEDLASAVSQLLKAAWRVQSEPLWVPASWVQ
Summary
Similarity
Belongs to the MAGUK family.
Uniprot
A0A194QRZ8
A0A194PLK2
A0A2W1BN52
A0A2H1VMY5
A0A2A4JGP1
H9ITY1
+ More
A0A1Y1MN50 A0A2J7PCD6 A0A139WLG4 E0W0X7 A0A1B6LBG4 A0A2P8ZJ44 A0A1B6EYV1 A0A1W4WK93 A0A1W4W8Y4 A0A1W4WK09 A0A2A3ESI0 A0A026WTD3 A0A1B6DRN7 A0A0K8VK16 A0A2J7PCD7 A0A158NL21 A0A0A1WUG2 A0A1B6D6R0 A0A232EHM8 A0A1A9YKL4 A0A1B0AM72 A0A224XM31 A0A182R6X1 A0A3L8DAP6 A0A1A9Z0N7 A0A1I8PC04 A0A023F3L4 A0A158NL20 A0A0L0BM97 F4WG01 A0A1B0GID1 T1HF44 A0A154NYM9 A0A084W5I3 A0A3B0JMV7 A0A1A9WNE4 N6W5E8 A0A182FMW4 A0A0Q9XIC1 A0A0M4E5D3 A0A0J9RB83 A0A0Q5VPQ4 A0A0N8NZZ8 Q28WS8 B7YZF0 A0A0Q9WNT9 A0A0Q9VXV8 B4H6B4 A0A1W4UGJ2 A0A0A9ZH83 A0A195BZW4 A0A0Q9XGG4 D6WID3 A0A0P8XLY2 A0A0J9RA78 B4HN43 B3NSM9 N6T3Q4 A0A195BH59 A1Z8G0 B4MK87 B4MFJ5 A0A0A9Z8D8 A0A1Y1MR77 A0A1W4U515 A0A182XM84 A0A182N8R9 A0A182QYM6 A0A182VSX1 A0A182JTC7 A0A212EGM5 A0A1B0FR55 A0A226E290 A0A1D2MHL8 A0A182P885 A0A336MAB8 B0W9U2 A0A0A9X2A7 A0A182GK60 A0A0A9ZDW1 A0A0P4WGQ6 A0A195F161 A0A151XBN5 A0A0P5JF50 A0A0P4YR38 A0A0P5CKL0 A0A0P6CIH3 A0A0P6FXE6 A0A0P6EUQ5 A0A0P6F9J6 A0A0P5IQ50 A0A195EFK9 A0A0P5V7N0 A0A0P5B6I2
A0A1Y1MN50 A0A2J7PCD6 A0A139WLG4 E0W0X7 A0A1B6LBG4 A0A2P8ZJ44 A0A1B6EYV1 A0A1W4WK93 A0A1W4W8Y4 A0A1W4WK09 A0A2A3ESI0 A0A026WTD3 A0A1B6DRN7 A0A0K8VK16 A0A2J7PCD7 A0A158NL21 A0A0A1WUG2 A0A1B6D6R0 A0A232EHM8 A0A1A9YKL4 A0A1B0AM72 A0A224XM31 A0A182R6X1 A0A3L8DAP6 A0A1A9Z0N7 A0A1I8PC04 A0A023F3L4 A0A158NL20 A0A0L0BM97 F4WG01 A0A1B0GID1 T1HF44 A0A154NYM9 A0A084W5I3 A0A3B0JMV7 A0A1A9WNE4 N6W5E8 A0A182FMW4 A0A0Q9XIC1 A0A0M4E5D3 A0A0J9RB83 A0A0Q5VPQ4 A0A0N8NZZ8 Q28WS8 B7YZF0 A0A0Q9WNT9 A0A0Q9VXV8 B4H6B4 A0A1W4UGJ2 A0A0A9ZH83 A0A195BZW4 A0A0Q9XGG4 D6WID3 A0A0P8XLY2 A0A0J9RA78 B4HN43 B3NSM9 N6T3Q4 A0A195BH59 A1Z8G0 B4MK87 B4MFJ5 A0A0A9Z8D8 A0A1Y1MR77 A0A1W4U515 A0A182XM84 A0A182N8R9 A0A182QYM6 A0A182VSX1 A0A182JTC7 A0A212EGM5 A0A1B0FR55 A0A226E290 A0A1D2MHL8 A0A182P885 A0A336MAB8 B0W9U2 A0A0A9X2A7 A0A182GK60 A0A0A9ZDW1 A0A0P4WGQ6 A0A195F161 A0A151XBN5 A0A0P5JF50 A0A0P4YR38 A0A0P5CKL0 A0A0P6CIH3 A0A0P6FXE6 A0A0P6EUQ5 A0A0P6F9J6 A0A0P5IQ50 A0A195EFK9 A0A0P5V7N0 A0A0P5B6I2
Pubmed
26354079
28756777
19121390
28004739
18362917
19820115
+ More
20566863 29403074 24508170 21347285 25830018 28648823 30249741 25474469 26108605 21719571 24438588 15632085 17994087 22936249 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25401762 23537049 22118469 27289101 26483478
20566863 29403074 24508170 21347285 25830018 28648823 30249741 25474469 26108605 21719571 24438588 15632085 17994087 22936249 18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25401762 23537049 22118469 27289101 26483478
EMBL
KQ461196
KPJ06316.1
KQ459605
KPI91985.1
KZ150032
PZC74707.1
+ More
ODYU01003429 SOQ42183.1 NWSH01001531 PCG70946.1 BABH01007151 GEZM01028066 JAV86488.1 NEVH01027062 PNF13999.1 KQ971321 KYB28746.1 DS235862 EEB19283.1 GEBQ01018962 JAT21015.1 PYGN01000040 PSN56526.1 GECZ01026926 JAS42843.1 KZ288187 PBC34715.1 KK107111 EZA58926.1 GEDC01008966 JAS28332.1 GDHF01013399 JAI38915.1 PNF14000.1 ADTU01019156 ADTU01019157 GBXI01011805 JAD02487.1 GEDC01015929 JAS21369.1 NNAY01004473 OXU17844.1 JXJN01000364 GFTR01007345 JAW09081.1 QOIP01000011 RLU16939.1 GBBI01002757 JAC15955.1 JRES01001669 KNC21048.1 GL888128 EGI66768.1 AJWK01013699 ACPB03000778 KQ434783 KZC04776.1 ATLV01020649 KE525304 KFB45477.1 OUUW01000001 SPP73971.1 CM000071 ENO01982.1 CH933808 KRG04049.1 CP012524 ALC41466.1 CM002911 KMY92919.1 CH954179 KQS62892.1 CH902619 KPU75723.1 KPU75724.1 EAL26589.2 AE013599 ACL83097.1 CH963846 KRF97593.1 CH940667 KRF77642.1 CH479213 EDW33338.1 GBHO01042968 GBHO01012562 GBHO01002074 JAG00636.1 JAG31042.1 JAG41530.1 KQ978501 KYM93466.1 KRG04050.1 EFA00746.2 KPU75721.1 KPU75722.1 KPU75725.1 KMY92921.1 CH480816 EDW47347.1 EDV56531.1 APGK01053668 KB741231 ENN72243.1 KQ976488 KYM83563.1 AAF58707.3 EDW72526.1 EDW57166.1 KRF77644.1 GBHO01042967 GBHO01002077 JAG00637.1 JAG41527.1 GEZM01028068 JAV86486.1 AXCN02001862 AGBW02015069 OWR40636.1 CCAG010014075 LNIX01000008 OXA50596.1 LJIJ01001219 ODM92476.1 UFQS01000752 UFQT01000752 SSX06623.1 SSX26970.1 DS231866 EDS40509.1 GBHO01029803 JAG13801.1 JXUM01012986 JXUM01012987 JXUM01012988 JXUM01012989 JXUM01012990 JXUM01012991 JXUM01012992 JXUM01012993 JXUM01012994 KQ560366 KXJ82790.1 GBHO01029791 GBHO01002076 JAG13813.1 JAG41528.1 GDRN01047779 JAI66707.1 KQ981866 KYN34121.1 KQ982320 KYQ57774.1 GDIP01250514 GDIP01250513 GDIP01225828 GDIQ01251049 GDIQ01247901 GDIQ01205160 GDIQ01203404 GDIP01044112 JAI72887.1 JAK48321.1 GDIP01225829 GDIQ01205161 GDIQ01203405 GDIP01071842 JAI97572.1 JAK46564.1 GDIP01169132 JAJ54270.1 GDIQ01090743 JAN03994.1 GDIQ01041518 JAN53219.1 GDIQ01165282 GDIQ01065059 JAN29678.1 GDIQ01063610 JAN31127.1 GDIQ01211954 JAK39771.1 KQ978957 KYN26951.1 GDIP01104491 JAL99223.1 GDIP01188826 JAJ34576.1
ODYU01003429 SOQ42183.1 NWSH01001531 PCG70946.1 BABH01007151 GEZM01028066 JAV86488.1 NEVH01027062 PNF13999.1 KQ971321 KYB28746.1 DS235862 EEB19283.1 GEBQ01018962 JAT21015.1 PYGN01000040 PSN56526.1 GECZ01026926 JAS42843.1 KZ288187 PBC34715.1 KK107111 EZA58926.1 GEDC01008966 JAS28332.1 GDHF01013399 JAI38915.1 PNF14000.1 ADTU01019156 ADTU01019157 GBXI01011805 JAD02487.1 GEDC01015929 JAS21369.1 NNAY01004473 OXU17844.1 JXJN01000364 GFTR01007345 JAW09081.1 QOIP01000011 RLU16939.1 GBBI01002757 JAC15955.1 JRES01001669 KNC21048.1 GL888128 EGI66768.1 AJWK01013699 ACPB03000778 KQ434783 KZC04776.1 ATLV01020649 KE525304 KFB45477.1 OUUW01000001 SPP73971.1 CM000071 ENO01982.1 CH933808 KRG04049.1 CP012524 ALC41466.1 CM002911 KMY92919.1 CH954179 KQS62892.1 CH902619 KPU75723.1 KPU75724.1 EAL26589.2 AE013599 ACL83097.1 CH963846 KRF97593.1 CH940667 KRF77642.1 CH479213 EDW33338.1 GBHO01042968 GBHO01012562 GBHO01002074 JAG00636.1 JAG31042.1 JAG41530.1 KQ978501 KYM93466.1 KRG04050.1 EFA00746.2 KPU75721.1 KPU75722.1 KPU75725.1 KMY92921.1 CH480816 EDW47347.1 EDV56531.1 APGK01053668 KB741231 ENN72243.1 KQ976488 KYM83563.1 AAF58707.3 EDW72526.1 EDW57166.1 KRF77644.1 GBHO01042967 GBHO01002077 JAG00637.1 JAG41527.1 GEZM01028068 JAV86486.1 AXCN02001862 AGBW02015069 OWR40636.1 CCAG010014075 LNIX01000008 OXA50596.1 LJIJ01001219 ODM92476.1 UFQS01000752 UFQT01000752 SSX06623.1 SSX26970.1 DS231866 EDS40509.1 GBHO01029803 JAG13801.1 JXUM01012986 JXUM01012987 JXUM01012988 JXUM01012989 JXUM01012990 JXUM01012991 JXUM01012992 JXUM01012993 JXUM01012994 KQ560366 KXJ82790.1 GBHO01029791 GBHO01002076 JAG13813.1 JAG41528.1 GDRN01047779 JAI66707.1 KQ981866 KYN34121.1 KQ982320 KYQ57774.1 GDIP01250514 GDIP01250513 GDIP01225828 GDIQ01251049 GDIQ01247901 GDIQ01205160 GDIQ01203404 GDIP01044112 JAI72887.1 JAK48321.1 GDIP01225829 GDIQ01205161 GDIQ01203405 GDIP01071842 JAI97572.1 JAK46564.1 GDIP01169132 JAJ54270.1 GDIQ01090743 JAN03994.1 GDIQ01041518 JAN53219.1 GDIQ01165282 GDIQ01065059 JAN29678.1 GDIQ01063610 JAN31127.1 GDIQ01211954 JAK39771.1 KQ978957 KYN26951.1 GDIP01104491 JAL99223.1 GDIP01188826 JAJ34576.1
Proteomes
UP000053240
UP000053268
UP000218220
UP000005204
UP000235965
UP000007266
+ More
UP000009046 UP000245037 UP000192223 UP000242457 UP000053097 UP000005205 UP000215335 UP000092443 UP000092460 UP000075900 UP000279307 UP000092445 UP000095300 UP000037069 UP000007755 UP000092461 UP000015103 UP000076502 UP000030765 UP000268350 UP000091820 UP000001819 UP000069272 UP000009192 UP000092553 UP000008711 UP000007801 UP000000803 UP000007798 UP000008792 UP000008744 UP000192221 UP000078542 UP000001292 UP000019118 UP000078540 UP000076407 UP000075884 UP000075886 UP000075920 UP000075881 UP000007151 UP000092444 UP000198287 UP000094527 UP000075885 UP000002320 UP000069940 UP000249989 UP000078541 UP000075809 UP000078492
UP000009046 UP000245037 UP000192223 UP000242457 UP000053097 UP000005205 UP000215335 UP000092443 UP000092460 UP000075900 UP000279307 UP000092445 UP000095300 UP000037069 UP000007755 UP000092461 UP000015103 UP000076502 UP000030765 UP000268350 UP000091820 UP000001819 UP000069272 UP000009192 UP000092553 UP000008711 UP000007801 UP000000803 UP000007798 UP000008792 UP000008744 UP000192221 UP000078542 UP000001292 UP000019118 UP000078540 UP000076407 UP000075884 UP000075886 UP000075920 UP000075881 UP000007151 UP000092444 UP000198287 UP000094527 UP000075885 UP000002320 UP000069940 UP000249989 UP000078541 UP000075809 UP000078492
Pfam
Interpro
IPR014775
L27_C
+ More
IPR004172 L27_dom
IPR008144 Guanylate_kin-like_dom
IPR001452 SH3_domain
IPR008145 GK/Ca_channel_bsu
IPR027417 P-loop_NTPase
IPR036028 SH3-like_dom_sf
IPR001478 PDZ
IPR036034 PDZ_sf
IPR020590 Guanylate_kinase_CS
IPR036892 L27_dom_sf
IPR041489 PDZ_6
IPR002110 Ankyrin_rpt
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
IPR004172 L27_dom
IPR008144 Guanylate_kin-like_dom
IPR001452 SH3_domain
IPR008145 GK/Ca_channel_bsu
IPR027417 P-loop_NTPase
IPR036028 SH3-like_dom_sf
IPR001478 PDZ
IPR036034 PDZ_sf
IPR020590 Guanylate_kinase_CS
IPR036892 L27_dom_sf
IPR041489 PDZ_6
IPR002110 Ankyrin_rpt
IPR036770 Ankyrin_rpt-contain_sf
IPR020683 Ankyrin_rpt-contain_dom
SUPFAM
Gene 3D
CDD
ProteinModelPortal
A0A194QRZ8
A0A194PLK2
A0A2W1BN52
A0A2H1VMY5
A0A2A4JGP1
H9ITY1
+ More
A0A1Y1MN50 A0A2J7PCD6 A0A139WLG4 E0W0X7 A0A1B6LBG4 A0A2P8ZJ44 A0A1B6EYV1 A0A1W4WK93 A0A1W4W8Y4 A0A1W4WK09 A0A2A3ESI0 A0A026WTD3 A0A1B6DRN7 A0A0K8VK16 A0A2J7PCD7 A0A158NL21 A0A0A1WUG2 A0A1B6D6R0 A0A232EHM8 A0A1A9YKL4 A0A1B0AM72 A0A224XM31 A0A182R6X1 A0A3L8DAP6 A0A1A9Z0N7 A0A1I8PC04 A0A023F3L4 A0A158NL20 A0A0L0BM97 F4WG01 A0A1B0GID1 T1HF44 A0A154NYM9 A0A084W5I3 A0A3B0JMV7 A0A1A9WNE4 N6W5E8 A0A182FMW4 A0A0Q9XIC1 A0A0M4E5D3 A0A0J9RB83 A0A0Q5VPQ4 A0A0N8NZZ8 Q28WS8 B7YZF0 A0A0Q9WNT9 A0A0Q9VXV8 B4H6B4 A0A1W4UGJ2 A0A0A9ZH83 A0A195BZW4 A0A0Q9XGG4 D6WID3 A0A0P8XLY2 A0A0J9RA78 B4HN43 B3NSM9 N6T3Q4 A0A195BH59 A1Z8G0 B4MK87 B4MFJ5 A0A0A9Z8D8 A0A1Y1MR77 A0A1W4U515 A0A182XM84 A0A182N8R9 A0A182QYM6 A0A182VSX1 A0A182JTC7 A0A212EGM5 A0A1B0FR55 A0A226E290 A0A1D2MHL8 A0A182P885 A0A336MAB8 B0W9U2 A0A0A9X2A7 A0A182GK60 A0A0A9ZDW1 A0A0P4WGQ6 A0A195F161 A0A151XBN5 A0A0P5JF50 A0A0P4YR38 A0A0P5CKL0 A0A0P6CIH3 A0A0P6FXE6 A0A0P6EUQ5 A0A0P6F9J6 A0A0P5IQ50 A0A195EFK9 A0A0P5V7N0 A0A0P5B6I2
A0A1Y1MN50 A0A2J7PCD6 A0A139WLG4 E0W0X7 A0A1B6LBG4 A0A2P8ZJ44 A0A1B6EYV1 A0A1W4WK93 A0A1W4W8Y4 A0A1W4WK09 A0A2A3ESI0 A0A026WTD3 A0A1B6DRN7 A0A0K8VK16 A0A2J7PCD7 A0A158NL21 A0A0A1WUG2 A0A1B6D6R0 A0A232EHM8 A0A1A9YKL4 A0A1B0AM72 A0A224XM31 A0A182R6X1 A0A3L8DAP6 A0A1A9Z0N7 A0A1I8PC04 A0A023F3L4 A0A158NL20 A0A0L0BM97 F4WG01 A0A1B0GID1 T1HF44 A0A154NYM9 A0A084W5I3 A0A3B0JMV7 A0A1A9WNE4 N6W5E8 A0A182FMW4 A0A0Q9XIC1 A0A0M4E5D3 A0A0J9RB83 A0A0Q5VPQ4 A0A0N8NZZ8 Q28WS8 B7YZF0 A0A0Q9WNT9 A0A0Q9VXV8 B4H6B4 A0A1W4UGJ2 A0A0A9ZH83 A0A195BZW4 A0A0Q9XGG4 D6WID3 A0A0P8XLY2 A0A0J9RA78 B4HN43 B3NSM9 N6T3Q4 A0A195BH59 A1Z8G0 B4MK87 B4MFJ5 A0A0A9Z8D8 A0A1Y1MR77 A0A1W4U515 A0A182XM84 A0A182N8R9 A0A182QYM6 A0A182VSX1 A0A182JTC7 A0A212EGM5 A0A1B0FR55 A0A226E290 A0A1D2MHL8 A0A182P885 A0A336MAB8 B0W9U2 A0A0A9X2A7 A0A182GK60 A0A0A9ZDW1 A0A0P4WGQ6 A0A195F161 A0A151XBN5 A0A0P5JF50 A0A0P4YR38 A0A0P5CKL0 A0A0P6CIH3 A0A0P6FXE6 A0A0P6EUQ5 A0A0P6F9J6 A0A0P5IQ50 A0A195EFK9 A0A0P5V7N0 A0A0P5B6I2
PDB
4WSI
E-value=7.14466e-45,
Score=457
Ontologies
GO
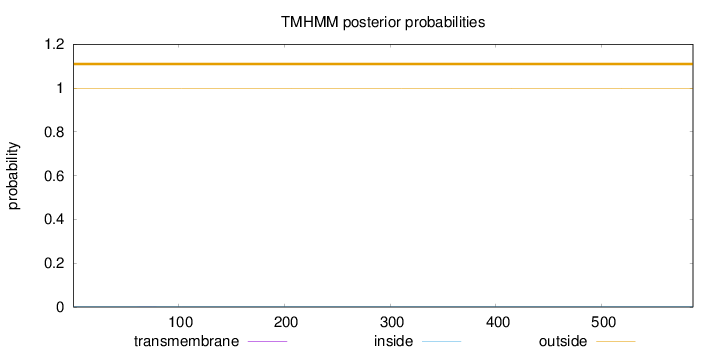
Topology
Length:
586
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00361
Exp number, first 60 AAs:
0
Total prob of N-in:
0.00116
outside
1 - 586
Population Genetic Test Statistics
Pi
27.210691
Theta
16.015182
Tajima's D
-1.274955
CLR
0.199456
CSRT
0.0917954102294885
Interpretation
Uncertain
