Gene
KWMTBOMO00362 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000710
Annotation
PREDICTED:_vesicle-associated_membrane_protein-associated_protein_B_isoform_X1_[Amyelois_transitella]
Location in the cell
Nuclear Reliability : 2.843
Sequence
CDS
ATGCCTAACCAAGTGCTAACTATCGAACCACAAAATGAATTAAAATTTAAAGTTAATTTCTCAGGGCTCTTTGAGCATGGTTGCACAACCTACATGAGACTAACGAACCCGACAAACGATACAGTGCTATTCAAGATCAAAACAACAGCACCAAAGAAATATTGCGTGCGTCCCAATTCTGGAGTTCTTGCGCCAAATTCGAAACAAGAAATCGCAATTACTCCGCAACCTGTATATTTGGACCCCAATGAAAACCACAAGCATAAATTTATGGTCCAAAGTGTAATAGCTCCAGAAGGAAAAACCAACATTGATCAAGTGTGGAAGGAGATAAGCCCCGAGCAACTCATGGACTACAAGCTAAAGTGTGTCTTTGAAACTCCGCGTGGCACTAATCTTAACGATTCTGGCGATAATGCAGCCCAGAACGATGTCACCAAGAAAAGAGTTGCAGTCGCAGAAGACGCTAAGCCTGCCGCTAAGGACGCCGTGAAAGATCTCATCCAGCATGAAGATAAAAATAAGGATGATGATCACCCTGTTGCATCAATAAACCTAAAAACGACTAATACCAAGTCTGAAAACGTTGAAAGCGATCTGCAAAAGGCGACTTTAGAAGTAATACATTTGAGAGAAGAGGAAAGCAAGTTAAGACATGAAAACCTGCAATTAAAAGAGGAGTTACTGCGCTTGCGTCAGTCGGCAGGCGGGGACGGGCGCGTCTCGCGTCCTCACTCTTACGGTCCGGAGAAGTCGACGCAAGTCACGATTATGCCGTGGGTCGTCACCGCAATATGCATGGCCATCATTGGCATCATTATTGTGCCTCGGGACGATACCATCTGGTGA
Protein
MPNQVLTIEPQNELKFKVNFSGLFEHGCTTYMRLTNPTNDTVLFKIKTTAPKKYCVRPNSGVLAPNSKQEIAITPQPVYLDPNENHKHKFMVQSVIAPEGKTNIDQVWKEISPEQLMDYKLKCVFETPRGTNLNDSGDNAAQNDVTKKRVAVAEDAKPAAKDAVKDLIQHEDKNKDDDHPVASINLKTTNTKSENVESDLQKATLEVIHLREEESKLRHENLQLKEELLRLRQSAGGDGRVSRPHSYGPEKSTQVTIMPWVVTAICMAIIGIIIVPRDDTIW
Summary
Uniprot
H9ITY0
A0A2A4JGV2
A0A2H1VV92
A0A2W1BI09
A0A2A4JGK5
A0A1E1WU19
+ More
A0A1E1WCR7 A0A1E1WT04 A0A194QL94 A0A212EGN0 A0A194PGY1 A0A0L7LR87 D6WHU6 A0A0T6BET0 A0A336LKW3 A0A336LPG0 A0A2J7REM4 A0A1W4WV52 A0A1B6H175 N6U4K9 A0A1B6KH63 A0A067RGG1 J3JW86 A0A1B6DRX8 A0A1B6HWG0 A0A0K8TP53 U5EXC9 Q17GP4 Q1HQV2 A0A1L8DWA6 A0A224XX18 A0A170XJH6 F0J957 A0A158NNR5 A0A1J1HJ45 A0A023FJE8 L7M5Q2 A0A131YTL7 A0A224ZBX3 A0A2P8YM85 B7QEF1 A0A151IAX8 A0A0C9QNX3 A0A2R7WA80 A0A1E1XTC7 E2A3F5 A0A0M8ZV57 A0A1W7R9A0 A0A2A3EJM4 V9II82 A0A087ZQD5 A0A293MWL6 W5JU07 E7D141 K7J935 E2C2M3
A0A1E1WCR7 A0A1E1WT04 A0A194QL94 A0A212EGN0 A0A194PGY1 A0A0L7LR87 D6WHU6 A0A0T6BET0 A0A336LKW3 A0A336LPG0 A0A2J7REM4 A0A1W4WV52 A0A1B6H175 N6U4K9 A0A1B6KH63 A0A067RGG1 J3JW86 A0A1B6DRX8 A0A1B6HWG0 A0A0K8TP53 U5EXC9 Q17GP4 Q1HQV2 A0A1L8DWA6 A0A224XX18 A0A170XJH6 F0J957 A0A158NNR5 A0A1J1HJ45 A0A023FJE8 L7M5Q2 A0A131YTL7 A0A224ZBX3 A0A2P8YM85 B7QEF1 A0A151IAX8 A0A0C9QNX3 A0A2R7WA80 A0A1E1XTC7 E2A3F5 A0A0M8ZV57 A0A1W7R9A0 A0A2A3EJM4 V9II82 A0A087ZQD5 A0A293MWL6 W5JU07 E7D141 K7J935 E2C2M3
Pubmed
EMBL
BABH01007148
NWSH01001531
PCG70948.1
ODYU01004640
SOQ44728.1
KZ150032
+ More
PZC74709.1 PCG70949.1 GDQN01011659 GDQN01000590 JAT79395.1 JAT90464.1 GDQN01006305 JAT84749.1 GDQN01000924 JAT90130.1 KQ461196 KPJ06333.1 AGBW02015069 OWR40634.1 KQ459605 KPI91964.1 JTDY01000271 KOB77977.1 KQ971321 EFA00056.1 LJIG01001111 KRT85829.1 UFQT01000041 SSX18682.1 SSX18681.1 NEVH01004959 PNF39292.1 GECZ01001336 JAS68433.1 APGK01042997 KB741011 KB632341 ENN75551.1 ERL92972.1 GEBQ01029169 JAT10808.1 KK852482 KDR22867.1 BT127504 AEE62466.1 GEDC01008861 JAS28437.1 GECU01028720 JAS78986.1 GDAI01001677 JAI15926.1 GANO01000129 JAB59742.1 CH477257 EAT45822.1 DQ440342 ABF18375.1 GFDF01003370 JAV10714.1 GFTR01003895 JAW12531.1 GEMB01004344 JAR98929.1 BK007408 DAA34113.1 ADTU01021735 ADTU01021736 CVRI01000004 CRK87436.1 GBBK01003352 JAC21130.1 GACK01005847 JAA59187.1 GEDV01006727 JAP81830.1 GFPF01013284 MAA24430.1 PYGN01000496 PSN45366.1 ABJB010804046 DS921134 EEC17233.1 KQ978169 KYM96598.1 GBYB01002332 JAG72099.1 KK854373 PTY15165.1 GFAA01000854 JAU02581.1 GL436428 EFN72019.1 KQ435867 KOX70253.1 GFAH01000667 JAV47722.1 KZ288223 PBC32013.1 JR048774 AEY60760.1 GFWV01019590 MAA44318.1 ADMH02000095 ETN67837.1 HQ005789 ADV40085.1 AAZX01003276 GL452187 EFN77801.1
PZC74709.1 PCG70949.1 GDQN01011659 GDQN01000590 JAT79395.1 JAT90464.1 GDQN01006305 JAT84749.1 GDQN01000924 JAT90130.1 KQ461196 KPJ06333.1 AGBW02015069 OWR40634.1 KQ459605 KPI91964.1 JTDY01000271 KOB77977.1 KQ971321 EFA00056.1 LJIG01001111 KRT85829.1 UFQT01000041 SSX18682.1 SSX18681.1 NEVH01004959 PNF39292.1 GECZ01001336 JAS68433.1 APGK01042997 KB741011 KB632341 ENN75551.1 ERL92972.1 GEBQ01029169 JAT10808.1 KK852482 KDR22867.1 BT127504 AEE62466.1 GEDC01008861 JAS28437.1 GECU01028720 JAS78986.1 GDAI01001677 JAI15926.1 GANO01000129 JAB59742.1 CH477257 EAT45822.1 DQ440342 ABF18375.1 GFDF01003370 JAV10714.1 GFTR01003895 JAW12531.1 GEMB01004344 JAR98929.1 BK007408 DAA34113.1 ADTU01021735 ADTU01021736 CVRI01000004 CRK87436.1 GBBK01003352 JAC21130.1 GACK01005847 JAA59187.1 GEDV01006727 JAP81830.1 GFPF01013284 MAA24430.1 PYGN01000496 PSN45366.1 ABJB010804046 DS921134 EEC17233.1 KQ978169 KYM96598.1 GBYB01002332 JAG72099.1 KK854373 PTY15165.1 GFAA01000854 JAU02581.1 GL436428 EFN72019.1 KQ435867 KOX70253.1 GFAH01000667 JAV47722.1 KZ288223 PBC32013.1 JR048774 AEY60760.1 GFWV01019590 MAA44318.1 ADMH02000095 ETN67837.1 HQ005789 ADV40085.1 AAZX01003276 GL452187 EFN77801.1
Proteomes
UP000005204
UP000218220
UP000053240
UP000007151
UP000053268
UP000037510
+ More
UP000007266 UP000235965 UP000192223 UP000019118 UP000030742 UP000027135 UP000008820 UP000005205 UP000183832 UP000245037 UP000001555 UP000078542 UP000000311 UP000053105 UP000242457 UP000005203 UP000000673 UP000002358 UP000008237
UP000007266 UP000235965 UP000192223 UP000019118 UP000030742 UP000027135 UP000008820 UP000005205 UP000183832 UP000245037 UP000001555 UP000078542 UP000000311 UP000053105 UP000242457 UP000005203 UP000000673 UP000002358 UP000008237
Pfam
PF00635 Motile_Sperm
Interpro
SUPFAM
SSF49354
SSF49354
Gene 3D
ProteinModelPortal
H9ITY0
A0A2A4JGV2
A0A2H1VV92
A0A2W1BI09
A0A2A4JGK5
A0A1E1WU19
+ More
A0A1E1WCR7 A0A1E1WT04 A0A194QL94 A0A212EGN0 A0A194PGY1 A0A0L7LR87 D6WHU6 A0A0T6BET0 A0A336LKW3 A0A336LPG0 A0A2J7REM4 A0A1W4WV52 A0A1B6H175 N6U4K9 A0A1B6KH63 A0A067RGG1 J3JW86 A0A1B6DRX8 A0A1B6HWG0 A0A0K8TP53 U5EXC9 Q17GP4 Q1HQV2 A0A1L8DWA6 A0A224XX18 A0A170XJH6 F0J957 A0A158NNR5 A0A1J1HJ45 A0A023FJE8 L7M5Q2 A0A131YTL7 A0A224ZBX3 A0A2P8YM85 B7QEF1 A0A151IAX8 A0A0C9QNX3 A0A2R7WA80 A0A1E1XTC7 E2A3F5 A0A0M8ZV57 A0A1W7R9A0 A0A2A3EJM4 V9II82 A0A087ZQD5 A0A293MWL6 W5JU07 E7D141 K7J935 E2C2M3
A0A1E1WCR7 A0A1E1WT04 A0A194QL94 A0A212EGN0 A0A194PGY1 A0A0L7LR87 D6WHU6 A0A0T6BET0 A0A336LKW3 A0A336LPG0 A0A2J7REM4 A0A1W4WV52 A0A1B6H175 N6U4K9 A0A1B6KH63 A0A067RGG1 J3JW86 A0A1B6DRX8 A0A1B6HWG0 A0A0K8TP53 U5EXC9 Q17GP4 Q1HQV2 A0A1L8DWA6 A0A224XX18 A0A170XJH6 F0J957 A0A158NNR5 A0A1J1HJ45 A0A023FJE8 L7M5Q2 A0A131YTL7 A0A224ZBX3 A0A2P8YM85 B7QEF1 A0A151IAX8 A0A0C9QNX3 A0A2R7WA80 A0A1E1XTC7 E2A3F5 A0A0M8ZV57 A0A1W7R9A0 A0A2A3EJM4 V9II82 A0A087ZQD5 A0A293MWL6 W5JU07 E7D141 K7J935 E2C2M3
PDB
3IKK
E-value=4.65042e-31,
Score=334
Ontologies
GO
PANTHER
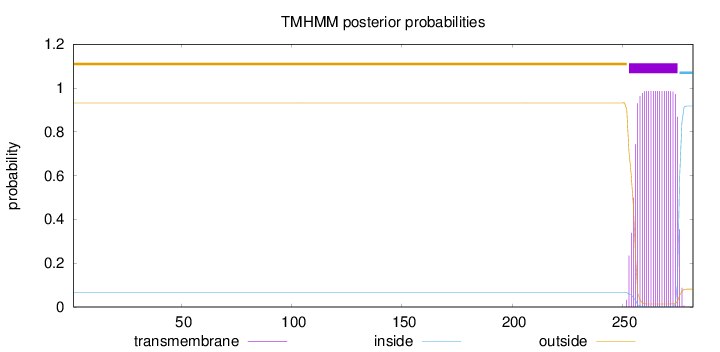
Topology
Length:
282
Number of predicted TMHs:
1
Exp number of AAs in TMHs:
20.80439
Exp number, first 60 AAs:
0.00099
Total prob of N-in:
0.06786
outside
1 - 252
TMhelix
253 - 275
inside
276 - 282
Population Genetic Test Statistics
Pi
17.252679
Theta
15.565357
Tajima's D
0.615028
CLR
0.15654
CSRT
0.560921953902305
Interpretation
Uncertain
