Gene
KWMTBOMO00361
Pre Gene Modal
BGIBMGA000518
Annotation
PREDICTED:_pyruvate_dehydrogenase_[acetyl-transferring]-phosphatase_1?_mitochondrial_[Amyelois_transitella]
Location in the cell
Mitochondrial Reliability : 2.891
Sequence
CDS
ATGAATAGTTCGAAACTGAATTTACTTTTGAACACAGTCAATACATATTTAAGATTAAACTCGGATGTAAAACAAACAATATTAAAAAAATACTTCAAAGAAAACTTCACATTATTCTGGAACACAACATCATCTAACACTCGCTATTTTCACTCTTCCTTGAAGGCAGCGCAAGTGCAAAAAGATGGGGTCACAGTATTACCTTCGCCTCAAGAAGTCACAACAATACTCCGAAAAAACGAATTCAACAAAGAATTCACAACTGGCTCCATTAAGTGCTACGATTCAAATCAACTCGCTTCAAACGACCCCATAGAGGATACTCGTTGTGAGGCACAATGTAGGATGACTTCAGGTTTGATTATGGGGGTGTTCGATGGTCATGGTGGACCTGCTTGTGCCCAAGTTATATCAAAGAGATTGGTTGATTACATAGCAGCCGCACTTCTGCCCATAGAAACATTGCACAAGTACATTCAGCAGGATCCTCGAGAAGCTTTGGTTGACACTTTTAATAATAGAGTTGAAATTTTACAGGAATTGAAAATTTTGTATGAGCGTAGTTTTAGACAATACTTAGCAGAGTTAGCTAATTTACCACATGAGGGTTATAACGTACCAACAGCTTTGGAACAAGCCATCATAAAAATTGACCAAGATTTGTCAAAGGAAGCAGTTGAGCCATATAAACTTAATCAGGTGATAAATCCCCAGACATTATCTGTAGCACTATCTGGTGCAGTGGCCTGTGTAACACATGTGGATGGGCCTAATCTGCATATTGCGAACGTTGGCGACTGTAATGCTGTTTTGGGTATTTTGACTGAAGACAAAGAGTGGATAGCGAAAAAAATAACAAAAGAACATAATTCCGAGAATTTCGATGAGTTGAAAAGAATTTGGAATGAGCATCCTGACAATGAGAGAAAAACTGTAATACGCAGAGATAGATTACTGGGTGAACTCGCTCCTTTACGTAGTTTGGGAGATTATAGATATAAATGGCCTATAGAGATTTTAATGAAAGTGGCAGTACCTTTAATTGGCCCAAAAGCTATACCTGCAAATTACCATACACCGCCTTATTTAACGGCAAAACCAGATTTATATTACCATAGATTGAGTACAAGGGATAAATTTTTAATTCTGGCTTCGGATGGATTATGGGATATGATGACGCCATTACAGGCAGTCAAGCTGGTGGGCGAGCATATGAAGGGAAAAGTTTTCTTTAACCCTTTGAAATTGCCCAGAAAGAATATTCAGCTGGGTGATATAAATGAATTACTCCTTCACAGGAAGGAAAGCTTGAAGAGTAAGCCAAAGGATAGAAATGCAGCCACACATTTAATACGTCATGCAATAGGTGGTACAGAATACGGCGTGGACCACTCCAGATTGGCACATTTGTTGAGTTTATCCCCGGATGTGAGTCGTATGTTTCGAGATGACATAACCGTTACGGTCATGTATTTTGATTCAGAGTTCTTAAGGCAATGTCCCGCATGA
Protein
MNSSKLNLLLNTVNTYLRLNSDVKQTILKKYFKENFTLFWNTTSSNTRYFHSSLKAAQVQKDGVTVLPSPQEVTTILRKNEFNKEFTTGSIKCYDSNQLASNDPIEDTRCEAQCRMTSGLIMGVFDGHGGPACAQVISKRLVDYIAAALLPIETLHKYIQQDPREALVDTFNNRVEILQELKILYERSFRQYLAELANLPHEGYNVPTALEQAIIKIDQDLSKEAVEPYKLNQVINPQTLSVALSGAVACVTHVDGPNLHIANVGDCNAVLGILTEDKEWIAKKITKEHNSENFDELKRIWNEHPDNERKTVIRRDRLLGELAPLRSLGDYRYKWPIEILMKVAVPLIGPKAIPANYHTPPYLTAKPDLYYHRLSTRDKFLILASDGLWDMMTPLQAVKLVGEHMKGKVFFNPLKLPRKNIQLGDINELLLHRKESLKSKPKDRNAATHLIRHAIGGTEYGVDHSRLAHLLSLSPDVSRMFRDDITVTVMYFDSEFLRQCPA
Summary
Cofactor
Mg(2+)
Similarity
Belongs to the PP2C family.
Uniprot
H9ITD8
A0A194PLI2
A0A0N1PFM7
A0A194QLC0
A0A2W1BPQ5
A0A2H1VV57
+ More
A0A2A4JG50 A0A212EGM2 A0A2J7REN1 A0A2P8YM86 J3JVN6 F4WY62 A0A067RQG2 A0A195CD95 A0A026WXB5 E9JBA2 A0A195EHI3 A0A195FDY9 A0A0J7KKM8 A0A2A3E402 A0A088A3M4 A0A151I411 E2BQY8 A0A158NT01 A0A151X6R6 A0A1Y1LPM3 A0A1B6IRR2 A0A0M9A883 A0A1B6G850 E2A1Y0 A0A1B6MU05 A0A139WKL9 A0A1Q3F3U6 Q16FI2 A0A0L7LRB1 A0A0L7QS58 W5JEK2 A0A182QAG2 A0A2M4ARG2 A0A1S4GY06 B0WYZ5 A0A182HA62 A0A182NTW9 Q7PNG1 A0A182IZZ7 A0A182IC96 A0A084W5U1 A0A182XLC4 A0A182UZB4 A0A1B6ED65 A0A1B6E0F1 A0A182K1R0 A0A182PUD9 A0A182M9Q4 A0A182W6I4 A0A182TJH6 A0A182Y3A0 K7J636 A0A154NYG5 A0A182T9Y8 A0A182RDN5 A0A1B0CI72 A0A1L8DM32 U5ESE6 A0A182UGV7 A0A0C9QQ18 A0A182FHI4 J9JMV4 A0A232F289 A0A2S2NLI0 A0A2H8TY53 A0A146KZY5 A0A0A9WX38 A0A1W4XES3 A0A1W4X3Z8 E0VZT0 A0A0N7Z9J7 T1HVJ0 A0A0V0GC59 A0A2R7WDX6 A0A336JYJ4 A0A1J1HWU8 T1IYH4 A0A2S2RA66 A0A1D2MAA9 A0A1B6CKV2 A0A182L0N8 A0A1S4EN31 B4MAE8 A0A087TIG8 B4L6C0 A0A224YZ29 A0A1A9Y8R4 A0A1B0BIV2 A0A131XKT2 A0A1W4V6G4 A0A0L0C961 A0A034WEB7 A0A240SWW7 B4JK29
A0A2A4JG50 A0A212EGM2 A0A2J7REN1 A0A2P8YM86 J3JVN6 F4WY62 A0A067RQG2 A0A195CD95 A0A026WXB5 E9JBA2 A0A195EHI3 A0A195FDY9 A0A0J7KKM8 A0A2A3E402 A0A088A3M4 A0A151I411 E2BQY8 A0A158NT01 A0A151X6R6 A0A1Y1LPM3 A0A1B6IRR2 A0A0M9A883 A0A1B6G850 E2A1Y0 A0A1B6MU05 A0A139WKL9 A0A1Q3F3U6 Q16FI2 A0A0L7LRB1 A0A0L7QS58 W5JEK2 A0A182QAG2 A0A2M4ARG2 A0A1S4GY06 B0WYZ5 A0A182HA62 A0A182NTW9 Q7PNG1 A0A182IZZ7 A0A182IC96 A0A084W5U1 A0A182XLC4 A0A182UZB4 A0A1B6ED65 A0A1B6E0F1 A0A182K1R0 A0A182PUD9 A0A182M9Q4 A0A182W6I4 A0A182TJH6 A0A182Y3A0 K7J636 A0A154NYG5 A0A182T9Y8 A0A182RDN5 A0A1B0CI72 A0A1L8DM32 U5ESE6 A0A182UGV7 A0A0C9QQ18 A0A182FHI4 J9JMV4 A0A232F289 A0A2S2NLI0 A0A2H8TY53 A0A146KZY5 A0A0A9WX38 A0A1W4XES3 A0A1W4X3Z8 E0VZT0 A0A0N7Z9J7 T1HVJ0 A0A0V0GC59 A0A2R7WDX6 A0A336JYJ4 A0A1J1HWU8 T1IYH4 A0A2S2RA66 A0A1D2MAA9 A0A1B6CKV2 A0A182L0N8 A0A1S4EN31 B4MAE8 A0A087TIG8 B4L6C0 A0A224YZ29 A0A1A9Y8R4 A0A1B0BIV2 A0A131XKT2 A0A1W4V6G4 A0A0L0C961 A0A034WEB7 A0A240SWW7 B4JK29
Pubmed
19121390
26354079
28756777
22118469
29403074
22516182
+ More
23537049 21719571 24845553 24508170 30249741 21282665 20798317 21347285 28004739 18362917 19820115 17510324 26227816 20920257 23761445 12364791 26483478 24438588 25244985 20075255 28648823 26823975 25401762 20566863 27129103 27289101 20966253 17994087 28797301 28049606 26108605 25348373
23537049 21719571 24845553 24508170 30249741 21282665 20798317 21347285 28004739 18362917 19820115 17510324 26227816 20920257 23761445 12364791 26483478 24438588 25244985 20075255 28648823 26823975 25401762 20566863 27129103 27289101 20966253 17994087 28797301 28049606 26108605 25348373
EMBL
BABH01007148
KQ459605
KPI91965.1
KQ459493
KPJ00081.1
KQ461196
+ More
KPJ06332.1 KZ150032 PZC74710.1 ODYU01004640 SOQ44727.1 NWSH01001531 PCG70951.1 AGBW02015069 OWR40633.1 NEVH01004959 PNF39289.1 PYGN01000496 PSN45368.1 APGK01053677 APGK01053678 APGK01053679 BT127304 KB741231 KB632387 AEE62266.1 ENN72249.1 ERL94350.1 GL888439 EGI60819.1 KK852482 KDR22865.1 KQ978023 KYM98048.1 KK107078 QOIP01000011 EZA60478.1 RLU16860.1 GL770938 EFZ09936.1 KQ978957 KYN27319.1 KQ981636 KYN38885.1 LBMM01006192 KMQ90837.1 KZ288403 PBC26224.1 KQ976472 KYM83988.1 GL449825 EFN81891.1 ADTU01025462 ADTU01025463 KQ982476 KYQ56052.1 GEZM01054558 JAV73556.1 GECU01018097 JAS89609.1 KQ435724 KOX78476.1 GECZ01011137 JAS58632.1 GL435856 EFN72556.1 GEBQ01000602 JAT39375.1 KQ971327 KYB28443.1 GFDL01012877 JAV22168.1 CH478433 EAT32992.1 JTDY01000271 KOB77985.1 KQ414758 KOC61480.1 ADMH02001732 ETN61254.1 AXCN02000807 GGFK01010038 MBW43359.1 AAAB01008964 DS232201 EDS37241.1 JXUM01030926 KQ560902 KXJ80420.1 EAA12222.4 APCN01000786 ATLV01020694 KE525305 KFB45585.1 GEDC01001425 JAS35873.1 GEDC01005909 JAS31389.1 AXCM01002012 KQ434783 KZC04715.1 AJWK01013066 GFDF01006649 JAV07435.1 GANO01003280 JAB56591.1 GBYB01005749 JAG75516.1 ABLF02021599 NNAY01001275 OXU24527.1 GGMR01005416 MBY18035.1 GFXV01007044 MBW18849.1 GDHC01016728 JAQ01901.1 GBHO01031615 GBHO01031613 JAG11989.1 JAG11991.1 DS235854 EEB18886.1 GDKW01000355 JAI56240.1 ACPB03010100 GECL01000568 JAP05556.1 KK854683 PTY17867.1 UFQS01000027 UFQT01000027 SSW97754.1 SSX18140.1 CVRI01000020 CRK90601.1 JH431683 GGMS01017728 MBY86931.1 LJIJ01002322 ODM89852.1 GEDC01023242 JAS14056.1 CH940655 EDW66207.1 KK115356 KFM64907.1 CH933812 EDW05916.1 GFPF01011049 MAA22195.1 JXJN01015182 GEFH01000467 JAP68114.1 JRES01000749 KNC28785.1 GAKP01006305 JAC52647.1 CCAG010017550 CH916370 EDV99931.1
KPJ06332.1 KZ150032 PZC74710.1 ODYU01004640 SOQ44727.1 NWSH01001531 PCG70951.1 AGBW02015069 OWR40633.1 NEVH01004959 PNF39289.1 PYGN01000496 PSN45368.1 APGK01053677 APGK01053678 APGK01053679 BT127304 KB741231 KB632387 AEE62266.1 ENN72249.1 ERL94350.1 GL888439 EGI60819.1 KK852482 KDR22865.1 KQ978023 KYM98048.1 KK107078 QOIP01000011 EZA60478.1 RLU16860.1 GL770938 EFZ09936.1 KQ978957 KYN27319.1 KQ981636 KYN38885.1 LBMM01006192 KMQ90837.1 KZ288403 PBC26224.1 KQ976472 KYM83988.1 GL449825 EFN81891.1 ADTU01025462 ADTU01025463 KQ982476 KYQ56052.1 GEZM01054558 JAV73556.1 GECU01018097 JAS89609.1 KQ435724 KOX78476.1 GECZ01011137 JAS58632.1 GL435856 EFN72556.1 GEBQ01000602 JAT39375.1 KQ971327 KYB28443.1 GFDL01012877 JAV22168.1 CH478433 EAT32992.1 JTDY01000271 KOB77985.1 KQ414758 KOC61480.1 ADMH02001732 ETN61254.1 AXCN02000807 GGFK01010038 MBW43359.1 AAAB01008964 DS232201 EDS37241.1 JXUM01030926 KQ560902 KXJ80420.1 EAA12222.4 APCN01000786 ATLV01020694 KE525305 KFB45585.1 GEDC01001425 JAS35873.1 GEDC01005909 JAS31389.1 AXCM01002012 KQ434783 KZC04715.1 AJWK01013066 GFDF01006649 JAV07435.1 GANO01003280 JAB56591.1 GBYB01005749 JAG75516.1 ABLF02021599 NNAY01001275 OXU24527.1 GGMR01005416 MBY18035.1 GFXV01007044 MBW18849.1 GDHC01016728 JAQ01901.1 GBHO01031615 GBHO01031613 JAG11989.1 JAG11991.1 DS235854 EEB18886.1 GDKW01000355 JAI56240.1 ACPB03010100 GECL01000568 JAP05556.1 KK854683 PTY17867.1 UFQS01000027 UFQT01000027 SSW97754.1 SSX18140.1 CVRI01000020 CRK90601.1 JH431683 GGMS01017728 MBY86931.1 LJIJ01002322 ODM89852.1 GEDC01023242 JAS14056.1 CH940655 EDW66207.1 KK115356 KFM64907.1 CH933812 EDW05916.1 GFPF01011049 MAA22195.1 JXJN01015182 GEFH01000467 JAP68114.1 JRES01000749 KNC28785.1 GAKP01006305 JAC52647.1 CCAG010017550 CH916370 EDV99931.1
Proteomes
UP000005204
UP000053268
UP000053240
UP000218220
UP000007151
UP000235965
+ More
UP000245037 UP000019118 UP000030742 UP000007755 UP000027135 UP000078542 UP000053097 UP000279307 UP000078492 UP000078541 UP000036403 UP000242457 UP000005203 UP000078540 UP000008237 UP000005205 UP000075809 UP000053105 UP000000311 UP000007266 UP000008820 UP000037510 UP000053825 UP000000673 UP000075886 UP000002320 UP000069940 UP000249989 UP000075884 UP000007062 UP000075880 UP000075840 UP000030765 UP000076407 UP000075903 UP000075881 UP000075885 UP000075883 UP000075920 UP000075902 UP000076408 UP000002358 UP000076502 UP000075901 UP000075900 UP000092461 UP000069272 UP000007819 UP000215335 UP000192223 UP000009046 UP000015103 UP000183832 UP000094527 UP000075882 UP000079169 UP000008792 UP000054359 UP000009192 UP000092443 UP000092460 UP000192221 UP000037069 UP000092444 UP000001070
UP000245037 UP000019118 UP000030742 UP000007755 UP000027135 UP000078542 UP000053097 UP000279307 UP000078492 UP000078541 UP000036403 UP000242457 UP000005203 UP000078540 UP000008237 UP000005205 UP000075809 UP000053105 UP000000311 UP000007266 UP000008820 UP000037510 UP000053825 UP000000673 UP000075886 UP000002320 UP000069940 UP000249989 UP000075884 UP000007062 UP000075880 UP000075840 UP000030765 UP000076407 UP000075903 UP000075881 UP000075885 UP000075883 UP000075920 UP000075902 UP000076408 UP000002358 UP000076502 UP000075901 UP000075900 UP000092461 UP000069272 UP000007819 UP000215335 UP000192223 UP000009046 UP000015103 UP000183832 UP000094527 UP000075882 UP000079169 UP000008792 UP000054359 UP000009192 UP000092443 UP000092460 UP000192221 UP000037069 UP000092444 UP000001070
PRIDE
Interpro
Gene 3D
ProteinModelPortal
H9ITD8
A0A194PLI2
A0A0N1PFM7
A0A194QLC0
A0A2W1BPQ5
A0A2H1VV57
+ More
A0A2A4JG50 A0A212EGM2 A0A2J7REN1 A0A2P8YM86 J3JVN6 F4WY62 A0A067RQG2 A0A195CD95 A0A026WXB5 E9JBA2 A0A195EHI3 A0A195FDY9 A0A0J7KKM8 A0A2A3E402 A0A088A3M4 A0A151I411 E2BQY8 A0A158NT01 A0A151X6R6 A0A1Y1LPM3 A0A1B6IRR2 A0A0M9A883 A0A1B6G850 E2A1Y0 A0A1B6MU05 A0A139WKL9 A0A1Q3F3U6 Q16FI2 A0A0L7LRB1 A0A0L7QS58 W5JEK2 A0A182QAG2 A0A2M4ARG2 A0A1S4GY06 B0WYZ5 A0A182HA62 A0A182NTW9 Q7PNG1 A0A182IZZ7 A0A182IC96 A0A084W5U1 A0A182XLC4 A0A182UZB4 A0A1B6ED65 A0A1B6E0F1 A0A182K1R0 A0A182PUD9 A0A182M9Q4 A0A182W6I4 A0A182TJH6 A0A182Y3A0 K7J636 A0A154NYG5 A0A182T9Y8 A0A182RDN5 A0A1B0CI72 A0A1L8DM32 U5ESE6 A0A182UGV7 A0A0C9QQ18 A0A182FHI4 J9JMV4 A0A232F289 A0A2S2NLI0 A0A2H8TY53 A0A146KZY5 A0A0A9WX38 A0A1W4XES3 A0A1W4X3Z8 E0VZT0 A0A0N7Z9J7 T1HVJ0 A0A0V0GC59 A0A2R7WDX6 A0A336JYJ4 A0A1J1HWU8 T1IYH4 A0A2S2RA66 A0A1D2MAA9 A0A1B6CKV2 A0A182L0N8 A0A1S4EN31 B4MAE8 A0A087TIG8 B4L6C0 A0A224YZ29 A0A1A9Y8R4 A0A1B0BIV2 A0A131XKT2 A0A1W4V6G4 A0A0L0C961 A0A034WEB7 A0A240SWW7 B4JK29
A0A2A4JG50 A0A212EGM2 A0A2J7REN1 A0A2P8YM86 J3JVN6 F4WY62 A0A067RQG2 A0A195CD95 A0A026WXB5 E9JBA2 A0A195EHI3 A0A195FDY9 A0A0J7KKM8 A0A2A3E402 A0A088A3M4 A0A151I411 E2BQY8 A0A158NT01 A0A151X6R6 A0A1Y1LPM3 A0A1B6IRR2 A0A0M9A883 A0A1B6G850 E2A1Y0 A0A1B6MU05 A0A139WKL9 A0A1Q3F3U6 Q16FI2 A0A0L7LRB1 A0A0L7QS58 W5JEK2 A0A182QAG2 A0A2M4ARG2 A0A1S4GY06 B0WYZ5 A0A182HA62 A0A182NTW9 Q7PNG1 A0A182IZZ7 A0A182IC96 A0A084W5U1 A0A182XLC4 A0A182UZB4 A0A1B6ED65 A0A1B6E0F1 A0A182K1R0 A0A182PUD9 A0A182M9Q4 A0A182W6I4 A0A182TJH6 A0A182Y3A0 K7J636 A0A154NYG5 A0A182T9Y8 A0A182RDN5 A0A1B0CI72 A0A1L8DM32 U5ESE6 A0A182UGV7 A0A0C9QQ18 A0A182FHI4 J9JMV4 A0A232F289 A0A2S2NLI0 A0A2H8TY53 A0A146KZY5 A0A0A9WX38 A0A1W4XES3 A0A1W4X3Z8 E0VZT0 A0A0N7Z9J7 T1HVJ0 A0A0V0GC59 A0A2R7WDX6 A0A336JYJ4 A0A1J1HWU8 T1IYH4 A0A2S2RA66 A0A1D2MAA9 A0A1B6CKV2 A0A182L0N8 A0A1S4EN31 B4MAE8 A0A087TIG8 B4L6C0 A0A224YZ29 A0A1A9Y8R4 A0A1B0BIV2 A0A131XKT2 A0A1W4V6G4 A0A0L0C961 A0A034WEB7 A0A240SWW7 B4JK29
PDB
3N3C
E-value=4.51867e-96,
Score=898
Ontologies
GO
PANTHER
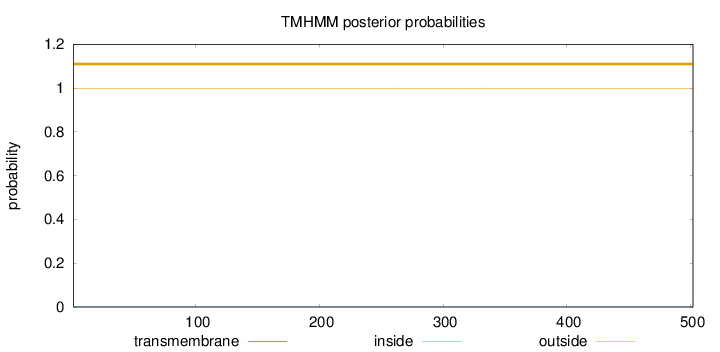
Topology
Length:
502
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.00358000000000001
Exp number, first 60 AAs:
0.00015
Total prob of N-in:
0.00068
outside
1 - 502
Population Genetic Test Statistics
Pi
14.349203
Theta
14.576493
Tajima's D
-0.549423
CLR
0.00024
CSRT
0.235138243087846
Interpretation
Uncertain
