Gene
KWMTBOMO00358
Pre Gene Modal
BGIBMGA000709
Annotation
PREDICTED:_frequenin-1_[Plutella_xylostella]
Full name
Frequenin-1
Alternative Name
d-FRQ
Location in the cell
Cytoplasmic Reliability : 3.073
Sequence
CDS
ATGAAGTTTGAACTTACGGAAAAAGAAATTCGGCTTTGGCACAAAGGATTTCTGAAGGACTGCCCCAACGGTCTTCTGACTGAGCAGGGCTTCATAAAGATCTACAAACAATTTTTCCCTCAAGGTGACCCGAGCAAGTTTGCGTCATTGGTGTTCAGAGTGTTTGACGAAAACAATGACGGGTCGATTGAGTTCGAAGAATTCATACGGGCCCTATCGGTGACGTCACGGGGCAATCTAGACGAGAAACTTCACTGGGCTTTCCGTCTCTACGACGTGGACAATGACGGTTACATCACTCGGGATGAAATGTATAACATAGTCGACGCGATCTATCAAATGGTGGGCCAGACCCCACAGCCTGAGGATGAGAACACTCCCCAGAAGCGCGTCGACAAGATCTTTGACCAGATGGACAAGAACCATGATGACAGGCTGACTCTTGAGGAATTCCGTGAGGGCAGCAAGGCCGACCCCCGCATTGTTCAGGCTCTGTCCCTTGGAGGCGATTAA
Protein
MKFELTEKEIRLWHKGFLKDCPNGLLTEQGFIKIYKQFFPQGDPSKFASLVFRVFDENNDGSIEFEEFIRALSVTSRGNLDEKLHWAFRLYDVDNDGYITRDEMYNIVDAIYQMVGQTPQPEDENTPQKRVDKIFDQMDKNHDDRLTLEEFREGSKADPRIVQALSLGGD
Summary
Description
Ca(2+)-dependent modulation of synaptic efficacy.
Miscellaneous
May bind 3 calcium ions via the second, third and fourth EF-hand.
Similarity
Belongs to the recoverin family.
Keywords
Calcium
Complete proteome
Lipoprotein
Membrane
Metal-binding
Myristate
Reference proteome
Repeat
Feature
chain Frequenin-1
Uniprot
A0A194PF22
A0A2A4JHS7
A0A2H1VVE6
A0A1Y1M0S7
A0A1S4FM07
A0A2M4CTA3
+ More
A0A2M4AZ97 A0A336KGG1 A0A182FJM8 A0A182RIL1 A0A182QRQ9 A0A182IQ91 A0A182W616 A0A084VGQ3 A0A1I8MYC6 A0A1I8Q235 A0A1W4XJE0 A0A3B0KMQ7 Q29G35 B4H7L2 B4MSM4 B4I6P7 B4JXS1 B3NTB4 A0A1W4USF5 B4L321 Q9VWX8 B3MWM1 B4M3A4 B4Q2V3 A0A2M4CVZ4 T1PAY1 A0A194QS13 A0A1B0G5A4 A0A1A9XWQ8 A0A1B0AUJ3 A0A2H8TKK4 A0A2S2NP06 A0A2S2QR16 A0A1B6CHL5 A0A1J1IWT3 A0A1B6KTB6 B4NTP0 K7IN11 A0A158NI29 A0A1I8NBS7 A0A0L0CK05 A0A3B0K158 B4H7L3 B4Q2V0 B3MWM3 B4MSM2 B4JXS4 B3NTB1 A0A1W4UQL6 M9PHW6 P37236 A0A0C9R5A7 A0A034VD77 A0A0K8WCY4 A0A2J7QMN2 A0A2W1BK99 A0A0K8WAE3 A0A034VCK0 A0A0A1WXY6 A0A0K8U2V0 A0A1B0FC88 D6WF01 B4NUL7 B4L2X8 A0A1A9X5J4 A0A3L8D951 W8AYK3 A0A069DWM2 E0VW56 A0A2M3ZK17 A0A164ZTL3 E9GDE5 E2AKT1 F4X100 A0A1B0A5D4 A0A0P6AW60 A0A026WV06 A0A1B6JMR2 Q9NB99 A0A0A7RNE9 A0A0J7LB52 A0A2M3ZDK3 A0A0A1WQ70 A0A154P8X1 H9ITX9 A0A1B0CME2 A0A1B0GQ97 B0XG62 W5JEC1 A0A182MPV7 A0A0M4FAT6 A0A067QLH6 W8BV80 A0A0M4ENJ4
A0A2M4AZ97 A0A336KGG1 A0A182FJM8 A0A182RIL1 A0A182QRQ9 A0A182IQ91 A0A182W616 A0A084VGQ3 A0A1I8MYC6 A0A1I8Q235 A0A1W4XJE0 A0A3B0KMQ7 Q29G35 B4H7L2 B4MSM4 B4I6P7 B4JXS1 B3NTB4 A0A1W4USF5 B4L321 Q9VWX8 B3MWM1 B4M3A4 B4Q2V3 A0A2M4CVZ4 T1PAY1 A0A194QS13 A0A1B0G5A4 A0A1A9XWQ8 A0A1B0AUJ3 A0A2H8TKK4 A0A2S2NP06 A0A2S2QR16 A0A1B6CHL5 A0A1J1IWT3 A0A1B6KTB6 B4NTP0 K7IN11 A0A158NI29 A0A1I8NBS7 A0A0L0CK05 A0A3B0K158 B4H7L3 B4Q2V0 B3MWM3 B4MSM2 B4JXS4 B3NTB1 A0A1W4UQL6 M9PHW6 P37236 A0A0C9R5A7 A0A034VD77 A0A0K8WCY4 A0A2J7QMN2 A0A2W1BK99 A0A0K8WAE3 A0A034VCK0 A0A0A1WXY6 A0A0K8U2V0 A0A1B0FC88 D6WF01 B4NUL7 B4L2X8 A0A1A9X5J4 A0A3L8D951 W8AYK3 A0A069DWM2 E0VW56 A0A2M3ZK17 A0A164ZTL3 E9GDE5 E2AKT1 F4X100 A0A1B0A5D4 A0A0P6AW60 A0A026WV06 A0A1B6JMR2 Q9NB99 A0A0A7RNE9 A0A0J7LB52 A0A2M3ZDK3 A0A0A1WQ70 A0A154P8X1 H9ITX9 A0A1B0CME2 A0A1B0GQ97 B0XG62 W5JEC1 A0A182MPV7 A0A0M4FAT6 A0A067QLH6 W8BV80 A0A0M4ENJ4
Pubmed
26354079
28004739
24438588
25315136
15632085
17994087
+ More
18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25074811 26109357 26109356 28119500 29966094 17550304 20075255 21347285 26108605 8101711 25348373 28756777 25830018 18362917 19820115 30249741 24495485 26334808 20566863 21292972 20798317 21719571 24508170 10512975 19121390 20920257 23761445 24845553
18057021 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 25074811 26109357 26109356 28119500 29966094 17550304 20075255 21347285 26108605 8101711 25348373 28756777 25830018 18362917 19820115 30249741 24495485 26334808 20566863 21292972 20798317 21719571 24508170 10512975 19121390 20920257 23761445 24845553
EMBL
KQ459605
KPI91966.1
NWSH01001531
PCG70952.1
ODYU01004640
SOQ44726.1
+ More
GEZM01044406 JAV78250.1 GGFL01004379 MBW68557.1 GGFK01012795 MBW46116.1 UFQS01000445 UFQT01000445 SSX03996.1 SSX24361.1 AXCN02002650 ATLV01013047 KE524836 KFB37147.1 OUUW01000011 SPP86411.1 CH379065 EAL31394.1 CH479218 EDW33823.1 CH963851 EDW75113.2 CH480823 EDW55995.1 CH916376 EDV95170.1 CH954180 EDV46565.1 CH933810 EDW06949.1 KRF93881.1 KRF93882.1 KRF93883.1 AE014298 BT133347 AAF48809.1 AAS65399.1 AAS65400.1 AFF57512.1 AGB95517.1 CH902625 EDV35006.1 KPU75387.1 CH940651 EDW65279.1 KRF82161.1 KRF82162.1 KRF82163.1 KRF82164.1 CM000162 EDX02687.1 KRK06927.1 KRK06928.1 GGFL01005334 MBW69512.1 KA645310 AFP59939.1 KQ461196 KPJ06331.1 CCAG010019898 CCAG010019899 JXJN01003666 GFXV01002860 MBW14665.1 GGMR01006286 MBY18905.1 GGMS01011013 MBY80216.1 GEDC01024341 JAS12957.1 CVRI01000063 CRL04560.1 GEBQ01025295 JAT14682.1 CH983253 EDX16309.1 ADTU01016194 ADTU01016195 ADTU01016196 ADTU01016197 ADTU01016198 JRES01000302 KNC32547.1 SPP86412.1 EDW33824.1 EDX02684.1 EDV35008.1 EDW75111.1 EDV95173.1 EDV46562.2 AGB95515.1 AGB95516.1 L08064 GBYB01007962 GBYB01007963 JAG77729.1 JAG77730.1 GAKP01019434 JAC39518.1 GDHF01028058 GDHF01025528 GDHF01018713 GDHF01003415 JAI24256.1 JAI26786.1 JAI33601.1 JAI48899.1 NEVH01013204 PNF29836.1 KZ150032 PZC74711.1 GDHF01004156 JAI48158.1 GAKP01019432 JAC39520.1 GBXI01010942 JAD03350.1 GDHF01033229 GDHF01031230 JAI19085.1 JAI21084.1 CCAG010010471 CCAG010010472 KQ971319 EFA00309.1 CH984109 EDX16664.1 EDW06945.2 QOIP01000011 RLU16438.1 GAMC01020623 GAMC01020621 GAMC01020619 GAMC01020618 GAMC01020617 JAB85936.1 GBGD01003160 JAC85729.1 DS235817 EEB17612.1 GGFM01008098 MBW28849.1 LRGB01000687 KZS16744.1 GL732540 EFX82492.1 GL440405 EFN65954.1 GL888520 EGI59860.1 GDIP01023554 JAM80161.1 KK107111 EZA58939.1 GECU01007182 JAT00525.1 AF260780 AAF74284.1 KM523324 AJA36971.1 LBMM01000056 KMR05137.1 GGFM01005865 MBW26616.1 GBXI01014299 GBXI01013729 GBXI01013701 GBXI01008116 GBXI01000820 GBXI01000748 JAC99992.1 JAD00563.1 JAD00591.1 JAD06176.1 JAD13472.1 JAD13544.1 KQ434846 KZC08376.1 BABH01007148 AJWK01018611 AJWK01018612 AJVK01006599 AJVK01006600 DS232995 EDS27280.1 ADMH02001734 ETN61250.1 AXCM01012451 CP012528 ALC49582.1 KK853183 KDR10143.1 GAMC01003463 JAC03093.1 ALC49584.1
GEZM01044406 JAV78250.1 GGFL01004379 MBW68557.1 GGFK01012795 MBW46116.1 UFQS01000445 UFQT01000445 SSX03996.1 SSX24361.1 AXCN02002650 ATLV01013047 KE524836 KFB37147.1 OUUW01000011 SPP86411.1 CH379065 EAL31394.1 CH479218 EDW33823.1 CH963851 EDW75113.2 CH480823 EDW55995.1 CH916376 EDV95170.1 CH954180 EDV46565.1 CH933810 EDW06949.1 KRF93881.1 KRF93882.1 KRF93883.1 AE014298 BT133347 AAF48809.1 AAS65399.1 AAS65400.1 AFF57512.1 AGB95517.1 CH902625 EDV35006.1 KPU75387.1 CH940651 EDW65279.1 KRF82161.1 KRF82162.1 KRF82163.1 KRF82164.1 CM000162 EDX02687.1 KRK06927.1 KRK06928.1 GGFL01005334 MBW69512.1 KA645310 AFP59939.1 KQ461196 KPJ06331.1 CCAG010019898 CCAG010019899 JXJN01003666 GFXV01002860 MBW14665.1 GGMR01006286 MBY18905.1 GGMS01011013 MBY80216.1 GEDC01024341 JAS12957.1 CVRI01000063 CRL04560.1 GEBQ01025295 JAT14682.1 CH983253 EDX16309.1 ADTU01016194 ADTU01016195 ADTU01016196 ADTU01016197 ADTU01016198 JRES01000302 KNC32547.1 SPP86412.1 EDW33824.1 EDX02684.1 EDV35008.1 EDW75111.1 EDV95173.1 EDV46562.2 AGB95515.1 AGB95516.1 L08064 GBYB01007962 GBYB01007963 JAG77729.1 JAG77730.1 GAKP01019434 JAC39518.1 GDHF01028058 GDHF01025528 GDHF01018713 GDHF01003415 JAI24256.1 JAI26786.1 JAI33601.1 JAI48899.1 NEVH01013204 PNF29836.1 KZ150032 PZC74711.1 GDHF01004156 JAI48158.1 GAKP01019432 JAC39520.1 GBXI01010942 JAD03350.1 GDHF01033229 GDHF01031230 JAI19085.1 JAI21084.1 CCAG010010471 CCAG010010472 KQ971319 EFA00309.1 CH984109 EDX16664.1 EDW06945.2 QOIP01000011 RLU16438.1 GAMC01020623 GAMC01020621 GAMC01020619 GAMC01020618 GAMC01020617 JAB85936.1 GBGD01003160 JAC85729.1 DS235817 EEB17612.1 GGFM01008098 MBW28849.1 LRGB01000687 KZS16744.1 GL732540 EFX82492.1 GL440405 EFN65954.1 GL888520 EGI59860.1 GDIP01023554 JAM80161.1 KK107111 EZA58939.1 GECU01007182 JAT00525.1 AF260780 AAF74284.1 KM523324 AJA36971.1 LBMM01000056 KMR05137.1 GGFM01005865 MBW26616.1 GBXI01014299 GBXI01013729 GBXI01013701 GBXI01008116 GBXI01000820 GBXI01000748 JAC99992.1 JAD00563.1 JAD00591.1 JAD06176.1 JAD13472.1 JAD13544.1 KQ434846 KZC08376.1 BABH01007148 AJWK01018611 AJWK01018612 AJVK01006599 AJVK01006600 DS232995 EDS27280.1 ADMH02001734 ETN61250.1 AXCM01012451 CP012528 ALC49582.1 KK853183 KDR10143.1 GAMC01003463 JAC03093.1 ALC49584.1
Proteomes
UP000053268
UP000218220
UP000069272
UP000075900
UP000075886
UP000075880
+ More
UP000075920 UP000030765 UP000095301 UP000095300 UP000192223 UP000268350 UP000001819 UP000008744 UP000007798 UP000001292 UP000001070 UP000008711 UP000192221 UP000009192 UP000000803 UP000007801 UP000008792 UP000002282 UP000053240 UP000092444 UP000092443 UP000092460 UP000183832 UP000000304 UP000002358 UP000005205 UP000037069 UP000235965 UP000007266 UP000091820 UP000279307 UP000009046 UP000076858 UP000000305 UP000000311 UP000007755 UP000092445 UP000053097 UP000036403 UP000076502 UP000005204 UP000092461 UP000092462 UP000002320 UP000000673 UP000075883 UP000092553 UP000027135
UP000075920 UP000030765 UP000095301 UP000095300 UP000192223 UP000268350 UP000001819 UP000008744 UP000007798 UP000001292 UP000001070 UP000008711 UP000192221 UP000009192 UP000000803 UP000007801 UP000008792 UP000002282 UP000053240 UP000092444 UP000092443 UP000092460 UP000183832 UP000000304 UP000002358 UP000005205 UP000037069 UP000235965 UP000007266 UP000091820 UP000279307 UP000009046 UP000076858 UP000000305 UP000000311 UP000007755 UP000092445 UP000053097 UP000036403 UP000076502 UP000005204 UP000092461 UP000092462 UP000002320 UP000000673 UP000075883 UP000092553 UP000027135
Interpro
SUPFAM
SSF47473
SSF47473
CDD
ProteinModelPortal
A0A194PF22
A0A2A4JHS7
A0A2H1VVE6
A0A1Y1M0S7
A0A1S4FM07
A0A2M4CTA3
+ More
A0A2M4AZ97 A0A336KGG1 A0A182FJM8 A0A182RIL1 A0A182QRQ9 A0A182IQ91 A0A182W616 A0A084VGQ3 A0A1I8MYC6 A0A1I8Q235 A0A1W4XJE0 A0A3B0KMQ7 Q29G35 B4H7L2 B4MSM4 B4I6P7 B4JXS1 B3NTB4 A0A1W4USF5 B4L321 Q9VWX8 B3MWM1 B4M3A4 B4Q2V3 A0A2M4CVZ4 T1PAY1 A0A194QS13 A0A1B0G5A4 A0A1A9XWQ8 A0A1B0AUJ3 A0A2H8TKK4 A0A2S2NP06 A0A2S2QR16 A0A1B6CHL5 A0A1J1IWT3 A0A1B6KTB6 B4NTP0 K7IN11 A0A158NI29 A0A1I8NBS7 A0A0L0CK05 A0A3B0K158 B4H7L3 B4Q2V0 B3MWM3 B4MSM2 B4JXS4 B3NTB1 A0A1W4UQL6 M9PHW6 P37236 A0A0C9R5A7 A0A034VD77 A0A0K8WCY4 A0A2J7QMN2 A0A2W1BK99 A0A0K8WAE3 A0A034VCK0 A0A0A1WXY6 A0A0K8U2V0 A0A1B0FC88 D6WF01 B4NUL7 B4L2X8 A0A1A9X5J4 A0A3L8D951 W8AYK3 A0A069DWM2 E0VW56 A0A2M3ZK17 A0A164ZTL3 E9GDE5 E2AKT1 F4X100 A0A1B0A5D4 A0A0P6AW60 A0A026WV06 A0A1B6JMR2 Q9NB99 A0A0A7RNE9 A0A0J7LB52 A0A2M3ZDK3 A0A0A1WQ70 A0A154P8X1 H9ITX9 A0A1B0CME2 A0A1B0GQ97 B0XG62 W5JEC1 A0A182MPV7 A0A0M4FAT6 A0A067QLH6 W8BV80 A0A0M4ENJ4
A0A2M4AZ97 A0A336KGG1 A0A182FJM8 A0A182RIL1 A0A182QRQ9 A0A182IQ91 A0A182W616 A0A084VGQ3 A0A1I8MYC6 A0A1I8Q235 A0A1W4XJE0 A0A3B0KMQ7 Q29G35 B4H7L2 B4MSM4 B4I6P7 B4JXS1 B3NTB4 A0A1W4USF5 B4L321 Q9VWX8 B3MWM1 B4M3A4 B4Q2V3 A0A2M4CVZ4 T1PAY1 A0A194QS13 A0A1B0G5A4 A0A1A9XWQ8 A0A1B0AUJ3 A0A2H8TKK4 A0A2S2NP06 A0A2S2QR16 A0A1B6CHL5 A0A1J1IWT3 A0A1B6KTB6 B4NTP0 K7IN11 A0A158NI29 A0A1I8NBS7 A0A0L0CK05 A0A3B0K158 B4H7L3 B4Q2V0 B3MWM3 B4MSM2 B4JXS4 B3NTB1 A0A1W4UQL6 M9PHW6 P37236 A0A0C9R5A7 A0A034VD77 A0A0K8WCY4 A0A2J7QMN2 A0A2W1BK99 A0A0K8WAE3 A0A034VCK0 A0A0A1WXY6 A0A0K8U2V0 A0A1B0FC88 D6WF01 B4NUL7 B4L2X8 A0A1A9X5J4 A0A3L8D951 W8AYK3 A0A069DWM2 E0VW56 A0A2M3ZK17 A0A164ZTL3 E9GDE5 E2AKT1 F4X100 A0A1B0A5D4 A0A0P6AW60 A0A026WV06 A0A1B6JMR2 Q9NB99 A0A0A7RNE9 A0A0J7LB52 A0A2M3ZDK3 A0A0A1WQ70 A0A154P8X1 H9ITX9 A0A1B0CME2 A0A1B0GQ97 B0XG62 W5JEC1 A0A182MPV7 A0A0M4FAT6 A0A067QLH6 W8BV80 A0A0M4ENJ4
PDB
6EPA
E-value=7.7688e-84,
Score=786
Ontologies
GO
PANTHER
Topology
Subcellular location
Membrane
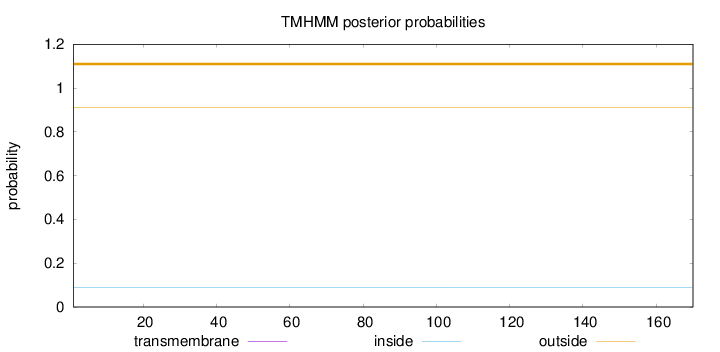
Length:
170
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0002
Exp number, first 60 AAs:
0.0002
Total prob of N-in:
0.09048
outside
1 - 170
Population Genetic Test Statistics
Pi
0
Theta
0
Tajima's D
0
CLR
0.44494
CSRT
0
Interpretation
Uncertain
