Gene
KWMTBOMO00349
Pre Gene Modal
BGIBMGA000705
Annotation
PREDICTED:_potassium_channel_subfamily_T_member_2_[Amyelois_transitella]
Full name
Potassium channel subfamily T member 1
Alternative Name
KCa4.1
Sequence like a calcium-activated potassium channel subunit
Sequence like a calcium-activated potassium channel subunit
Location in the cell
PlasmaMembrane Reliability : 4.447
Sequence
CDS
ATGGCAGGTGTCAGTAGATATAGTTCATACAATTGTTTGACTGGCTGGGACAGCTATGACAAGATACCCAGAGTAAGGGTGGAGTTCTACGTCAACGAGAACACTTTTAAGGAAAGACTGCAATTATATTTTATTAAAAATCAACGATCGAGTTTACGCATTAGGATAGCGAATCTATTTTTCAAACTTTTGACCTGCGTGCTGTATATATTTCGTGTGTGTACTGAAGGAGATCCTATAGCTGCGACTTGTTATGGATGTAAGCTTGGAAACAAGACTGACTTCCAACATTCTGCCAATTTGACGGAAGAAGAGTTCCAAGAGCACCCGTTAATTAATTGGGACGCAATAATATGGGTCAACCGACCTTTATATCTATGGATAGTGCAAGTACTCCTGGCATTGGTGTCTTTCACCGAAGCATTATTACTTGCCTACTTGGGCTATAAGGGCAACATTTGGCAGCAAGTGTTATCATTCCACTTCATCTTGGAAATGGTGAACACTATACCCTTTGCGTTAACCCTGACATTTCCTCCATTACGAAATCTATTTATACCGGTCTTTCTGAATTGTTGGCTCGCAAAAAGATCACTTGAAAATATGTTTAACGATTTACATCGAGCTATGCAAAAGTCGCAATCAGCTTTATCACAACAATTAATGATATTATGCGTTACGCTACTCTGTCTCGTCTTTACTAGTGTCTGCGGTATTCAACACTTTCAACGAGCTGGACATCGACACCTTAATCTTTTCCAAGCCACTTATTTTGTCGTAGTGACATTTTCAACAGTGGGCTATGGCGATTTTGTTCCGGACATATGGCCCTCTCAATTGTTCATGGTTGTCATGATTGGAGTTGCGCTGGTTGTTCTTCCAACACAGTTTGAACAATTGGCCTTTACTTGGATGGAAAGACAGAAATTGGGAGGATCATATTCTTCTCACCGAGCACAGTCTGAAAAACATGTAGTAGTATGTTCAACAACTTTACATGCCGACACTATCATGGACTTCTTGAACGAATTCTACGCACATCCACTACTTCAAGACTATTATGTTGTATTGCTGTCGCCGATGGAACTCGATACTACTATGCGTATGATTCTGCAGGTACCGATTTGGGCTCAACGTGTAATTTACGTTCAAGGCTCATGCCTAAAAGATACGGATTTGATTAGAGCTCGTATGAACGAAGCTGAAGCCTGTTTCATCTTGGCCGCCAGAAATTATGCCGACAAAACCGCTGCAGACGAACATACAATTTTACGATCATGGGCCGTTAAAGATTTCGCACCAAAAGTTGCACAATATGTACAAATATTTAGACCAGAAAATAAGCTACATGTAAAATTTGCTGAATTCGTGGTATGTGAAGATGAATTTAAATATGCTTTGTTGGCTAACAACTGTACCTGTCCAGGAGCGTCAACTTTAGTAACTCTCCTGCTTCACACAAGCAGAGGACAAGAAGGACAACAATCGCCTGAAGAATGGCACCGCCTTTATGGAAAATGTTCCGGAAATGAAATATACCACATCGTTCTGGGAGACAGCAGATTTTTCGGAGAATATGAAGGAAAAAGTTTTACTTACGCTAGTTTTCACTCACACCGAAAGTATGGAGTCGCTTTAGTTGGTGTTCGGCCCGCTGAATTACCTGAATTTTATGAAGATACAATATTGCTTAATCCGGGACCTCGGCATATAATGAAAAGCACCGATACGTGTTATTATATGAGTATAACAAAAGAAGAGAATTCCGCTTTTGTTGTTTCTGATAAACAGGCTGTACCAAACCCTACGATACCAAAAGACCAAACAGCATGCAGCTTCCTTTCAAACGAGAGGAATCCAGACGAAGGCAGTACAAGCCAGCAGAAGAAACCAGACGCTAAATTAGCCGATCGCTCTACCTCCGAATCAAGGACCTGCTTAAAAGAATCACCAGGAATCGAAAGTGTTGTTGACAGTCATTTACTTTTACCGAAACCTGAAACCAGCAATTTCTTAAGTCCGGACACACTGAGTCATCGTGGAGGAAATAGCAGGAGGCCATCGATACTTCCTGTTCCAGATATGGTCACCAGTAGTTTAACTTTAACAAATGACAATCAAGATGAAGAAGCTGAAATTGATGAGAGCGAAGATGAGCTAGATGACGATGTTCCTTGGCGATCCCCATCAGAAAAAATTGCAGATGACTTCACTACAAGCAGTTTCTCCTCGCTGGCTGCCAGTGATAACGCACACAGCGCTTGGCGCAACGATTTTGCAAGAATCGTCAAAGGATTCCCGCCTGTGTCACCTTTTATTGGAGTCAGCCCAACTCTCTGTTTTCTGCTAAAGGAAAAGAAACCTCTTTGCTGCTTACAACTGGCCCAAGTCTGTGAACACTGCGCCTATAGAAATGCCAAGGAATATCAATGGCAAAATAAAACGATTATTCTTGCTGCCGACTATGCTTCTAATGGAATTTACAATTTTATTATTCCGCTAAGAGCTCATTTTCGGTCTAAAACAACTTTAAACCCTATTATTTTGCTTCTCGAGCGACGGCCAGATGTTGCATTTCTAGATGCCATATCTTATTTCCCTCTGGTTTATTGGATGTTGGGCACTATTGATTGCTTGGATGATCTGCTTCGAGCTGGAATAACATTAGCTGAAAACGTTGTAGTTGTGAACAAAGAGCTATCAAATTCTGCTGAGGAAGATAGCCTTGCTGATTGCAATACAATTGTAGCAGTACAGACAATGTTCAAGTTCTTCCCTTCAATAAGGTCCATAACAGAACTATCTCAATCATCAAATATGAGATTTATGCAATTCCGTGCTCACGATAAATATGCTTTACATCTTTCTAAGATGGAAAAGCGAGAAAAAGAAAGGGGCTCACACATTTCCTATATGTTTCGATTGCCTTTCGCGGCCGGATCAGTTTTCTCTGCCTCAATGTTGGACACGTTACTTTATCAAGCCTTTGTTAAAGATTACATGATTACTTTTGTGCGACTATTATTGGGAATAGATCAGGCGCCCGGATCTGGATTTTTAGCGTCGATAAAAATTACCAAAGAAGATATGTGGATTCGTACCTATGGTCGTTTATACCAAAAACTATGTTCGACCTCATGTGAAATCCCAATTGGGATATATCGCACACAAGATACAAGCATGGCCGATGCATCTCACCACCAGGAGGAGCTTGATGGAGGAGATTTCCCGGCGTGCGGGCGTCAAGAGGCAACTAGCTGCCTTGGTGGTTGCTACGGAGAACGCACGCCGGTGTACTCTACAAGTCTGGCAGATGAAGCTCGCGACAATCACATTCAGCAAATTGAAAGAGCTGAAATAGCCAACTTGGTTCGTTCCAGGATGGAATCTCTAAATCTAACTGGGGTAGATTACGATGATGTAAGCGAAAAACGAAATAGTTTATCTTATGTCATAATCAATCCAAGCTGCGACTTAAATTTGCAAGAAGGAGACATCATTTATCTTGTGCGCCCTTCTCCATTTTCTGCACAAAAAACATTTGAAAGGCATAACAGTCGTCGAAAATCAAATATAAGCTTCTGTTCTCAAGCCTTAATCCAAGCGCAGGGAGGGAGCAGACGAGGTTCAGGAATGGCAGCAACTTTTGCTCAATCACGGGCTCCACCTCTTTCCACTACCAAGGCTAATTCGTTGTCTTTACCCGATAGCCCAACAATCATTACAGATTTCAGAGGTCGCTCTAATTCACTGCGTGTAGTGGATGATATTTTACTACGACGATCAAATTCTTTACGTCAGGGCATAAGCGGAACAGGATCCCGACGTCGCAAAAGCAGTTTAGAAGAAATAGGCATATCGCACTTCAATTCCTTGCTTCAACATCAGCAGCAACAGCAGGAGCAAGATGCTATAAAATTAGGACTAAGCAGCATTGGCCTAAGAATTACACCACCGGAAGAAACTCCTCGAATGGCTTTAGAGCCATTCCAGGACATAGCAGCAGCACTACCATCTACATCAGGTGGATCCACCATGCCAGTCAATGTAACTCCTGACCCACAACATCTTCAGGGTACAATCGTGTGA
Protein
MAGVSRYSSYNCLTGWDSYDKIPRVRVEFYVNENTFKERLQLYFIKNQRSSLRIRIANLFFKLLTCVLYIFRVCTEGDPIAATCYGCKLGNKTDFQHSANLTEEEFQEHPLINWDAIIWVNRPLYLWIVQVLLALVSFTEALLLAYLGYKGNIWQQVLSFHFILEMVNTIPFALTLTFPPLRNLFIPVFLNCWLAKRSLENMFNDLHRAMQKSQSALSQQLMILCVTLLCLVFTSVCGIQHFQRAGHRHLNLFQATYFVVVTFSTVGYGDFVPDIWPSQLFMVVMIGVALVVLPTQFEQLAFTWMERQKLGGSYSSHRAQSEKHVVVCSTTLHADTIMDFLNEFYAHPLLQDYYVVLLSPMELDTTMRMILQVPIWAQRVIYVQGSCLKDTDLIRARMNEAEACFILAARNYADKTAADEHTILRSWAVKDFAPKVAQYVQIFRPENKLHVKFAEFVVCEDEFKYALLANNCTCPGASTLVTLLLHTSRGQEGQQSPEEWHRLYGKCSGNEIYHIVLGDSRFFGEYEGKSFTYASFHSHRKYGVALVGVRPAELPEFYEDTILLNPGPRHIMKSTDTCYYMSITKEENSAFVVSDKQAVPNPTIPKDQTACSFLSNERNPDEGSTSQQKKPDAKLADRSTSESRTCLKESPGIESVVDSHLLLPKPETSNFLSPDTLSHRGGNSRRPSILPVPDMVTSSLTLTNDNQDEEAEIDESEDELDDDVPWRSPSEKIADDFTTSSFSSLAASDNAHSAWRNDFARIVKGFPPVSPFIGVSPTLCFLLKEKKPLCCLQLAQVCEHCAYRNAKEYQWQNKTIILAADYASNGIYNFIIPLRAHFRSKTTLNPIILLLERRPDVAFLDAISYFPLVYWMLGTIDCLDDLLRAGITLAENVVVVNKELSNSAEEDSLADCNTIVAVQTMFKFFPSIRSITELSQSSNMRFMQFRAHDKYALHLSKMEKREKERGSHISYMFRLPFAAGSVFSASMLDTLLYQAFVKDYMITFVRLLLGIDQAPGSGFLASIKITKEDMWIRTYGRLYQKLCSTSCEIPIGIYRTQDTSMADASHHQEELDGGDFPACGRQEATSCLGGCYGERTPVYSTSLADEARDNHIQQIERAEIANLVRSRMESLNLTGVDYDDVSEKRNSLSYVIINPSCDLNLQEGDIIYLVRPSPFSAQKTFERHNSRRKSNISFCSQALIQAQGGSRRGSGMAATFAQSRAPPLSTTKANSLSLPDSPTIITDFRGRSNSLRVVDDILLRRSNSLRQGISGTGSRRRKSSLEEIGISHFNSLLQHQQQQQEQDAIKLGLSSIGLRITPPEETPRMALEPFQDIAAALPSTSGGSTMPVNVTPDPQHLQGTIV
Summary
Description
Outwardly rectifying potassium channel subunit that may coassemble with other Slo-type channel subunits. Activated by high intracellular sodium or chloride levels. Activated upon stimulation of G-protein coupled receptors, such as CHRM1 and GRIA1. May be regulated by calcium in the absence of sodium ions (in vitro) (By similarity).
Generates outwardly rectifying currents that are suppressed by elevation of intracellular calcium.
Generates outwardly rectifying currents that are suppressed by elevation of intracellular calcium.
Subunit
Interacts (via C-terminus) with FMR1; this interaction alters gating properties of KCNT1 (PubMed:20512134). Interacts with CRBN via its cytoplasmic C-terminus (By similarity).
Similarity
Belongs to the potassium channel family. Calcium-activated (TC 1.A.1.3) subfamily. KCa4.1/KCNT1 sub-subfamily.
Keywords
Alternative splicing
Calcium
Cell membrane
Complete proteome
Disease mutation
Epilepsy
Glycoprotein
Ion channel
Ion transport
Membrane
Phosphoprotein
Potassium
Potassium channel
Potassium transport
Reference proteome
Transmembrane
Transmembrane helix
Transport
3D-structure
Feature
chain Potassium channel subfamily T member 1
splice variant In isoform 2 and isoform 3.
sequence variant In EIEE14; dbSNP:rs587777264.
splice variant In isoform 2 and isoform 3.
sequence variant In EIEE14; dbSNP:rs587777264.
Uniprot
A0A212EY19
A0A194QWH0
A0A194PY46
A0A139WLY1
A0A139WM47
A7UR92
+ More
A0A182IR49 A0A3B0JKV1 A0A1W4XHF4 A0A3B0JLH8 J9K6K2 A0A336LSZ5 A0A336LTP8 A0A336LUK8 A0A336LQC2 A0A336LQ39 A0A336LPP9 A0A1J1ITI9 A0A1Y1NBK9 A0A336LPT4 A0A1Y1NE12 A0A2S2NZ98 A0A336LPM5 A0A1J1IRM2 E0W0S8 A0A336M243 A0A0P5ZTV5 A0A162QJ56 A0A0P5ZX15 A0A0P6CR92 A0A0P5B3Q8 A0A0P5XMA2 A0A0P6A468 N6UDZ1 A0A0P5DU19 E9HEH7 A0A0P5WSA0 A0A0P5AC24 A0A0L7QTU4 A0A0P6IQA5 A0A0P6ATN0 A0A1B0CHQ2 U4TUR2 U4UE65 A0A1C9TA78 K1PSX5 A0A0L8G7B1 A0A0L8G6V3 A0A3B3I015 A0A336LPL1 A0A267EGF8 A0A3B4VM99 A0A3B4ZIJ9 A0A336LTT0 A0A2K5RPJ3 I3N3P6 A0A2I3S2J6 A0A2K5CTR7 Q5JUK3-3 H0WYR9 A0A2J8UIJ8 A0A2K6GK25 A0A2K6K4N9 A0A2K6NPW6 A0A146VBH1 A0A0K2GMW5 A0A3P8XE41 A0A3P9KR47 A0A3B4VM72 A0A3P8XB84 E1BW18 Q8QFV0 A0A3P9IIP2 A0A2K6DE59 A0A1W5ACB6 A0A3B4VIR6 A0A2K5RPF5 A0A2K5CTT5 A0A2I3T3B9 A0A3P8XB73 A0A2I2YBE5 A0A2K5RPK7 A0A287D7E5 A0A2J8NA34 A0A3B3RM10 A0A2K5CTS9 Q5JUK3-2 A0A2K6K4Q0 A0A0D9SFC8 A0A2I2YX38 G3QR87 A0A2J8UIP1 A0A2J8UIN6 A0A3B4EPN9 A0A2K5CTS6 A0A2K6GK47 A0A3B3RND9 A0A2K6K4M7 A0A2K6NPX6 A0A146VC13
A0A182IR49 A0A3B0JKV1 A0A1W4XHF4 A0A3B0JLH8 J9K6K2 A0A336LSZ5 A0A336LTP8 A0A336LUK8 A0A336LQC2 A0A336LQ39 A0A336LPP9 A0A1J1ITI9 A0A1Y1NBK9 A0A336LPT4 A0A1Y1NE12 A0A2S2NZ98 A0A336LPM5 A0A1J1IRM2 E0W0S8 A0A336M243 A0A0P5ZTV5 A0A162QJ56 A0A0P5ZX15 A0A0P6CR92 A0A0P5B3Q8 A0A0P5XMA2 A0A0P6A468 N6UDZ1 A0A0P5DU19 E9HEH7 A0A0P5WSA0 A0A0P5AC24 A0A0L7QTU4 A0A0P6IQA5 A0A0P6ATN0 A0A1B0CHQ2 U4TUR2 U4UE65 A0A1C9TA78 K1PSX5 A0A0L8G7B1 A0A0L8G6V3 A0A3B3I015 A0A336LPL1 A0A267EGF8 A0A3B4VM99 A0A3B4ZIJ9 A0A336LTT0 A0A2K5RPJ3 I3N3P6 A0A2I3S2J6 A0A2K5CTR7 Q5JUK3-3 H0WYR9 A0A2J8UIJ8 A0A2K6GK25 A0A2K6K4N9 A0A2K6NPW6 A0A146VBH1 A0A0K2GMW5 A0A3P8XE41 A0A3P9KR47 A0A3B4VM72 A0A3P8XB84 E1BW18 Q8QFV0 A0A3P9IIP2 A0A2K6DE59 A0A1W5ACB6 A0A3B4VIR6 A0A2K5RPF5 A0A2K5CTT5 A0A2I3T3B9 A0A3P8XB73 A0A2I2YBE5 A0A2K5RPK7 A0A287D7E5 A0A2J8NA34 A0A3B3RM10 A0A2K5CTS9 Q5JUK3-2 A0A2K6K4Q0 A0A0D9SFC8 A0A2I2YX38 G3QR87 A0A2J8UIP1 A0A2J8UIN6 A0A3B4EPN9 A0A2K5CTS6 A0A2K6GK47 A0A3B3RND9 A0A2K6K4M7 A0A2K6NPX6 A0A146VC13
Pubmed
EMBL
AGBW02011631
OWR46385.1
KQ461108
KPJ09310.1
KQ459594
KPI96070.1
+ More
KQ971314 KYB29019.1 KYB29020.1 AAAB01008807 EDO64534.2 OUUW01000001 SPP74099.1 SPP74096.1 ABLF02039742 ABLF02039754 ABLF02047215 UFQT01000105 SSX20041.1 SSX20043.1 SSX20037.1 SSX20045.1 SSX20040.1 SSX20036.1 CVRI01000058 CRL02892.1 GEZM01007528 JAV95109.1 SSX20042.1 GEZM01007527 JAV95110.1 GGMR01009773 MBY22392.1 SSX20044.1 CRL02893.1 DS235862 EEB19234.1 SSX20038.1 GDIP01039686 JAM64029.1 LRGB01000327 KZS19731.1 GDIP01038538 JAM65177.1 GDIP01000470 JAN03246.1 GDIP01190159 JAJ33243.1 GDIP01082322 JAM21393.1 GDIP01035246 JAM68469.1 APGK01026655 APGK01026656 APGK01026657 APGK01026658 APGK01026659 KB740635 ENN79925.1 GDIP01152820 JAJ70582.1 GL732629 EFX69876.1 GDIP01082323 JAM21392.1 GDIP01202063 JAJ21339.1 KQ414736 KOC62087.1 GDIQ01001674 JAN93063.1 GDIP01038537 JAM65178.1 AJWK01012580 AJWK01012581 AJWK01012582 AJWK01012583 AJWK01012584 AJWK01012585 KB630556 ERL83673.1 KB632107 ERL88866.1 KU681442 AOR07222.1 JH818227 EKC24758.1 KQ423495 KOF72763.1 KOF72762.1 SSX20034.1 NIVC01002133 PAA60613.1 SSX20039.1 AGTP01090746 AGTP01090747 AGTP01090748 AACZ04057952 NBAG03000232 PNI68601.1 AK127272 AL158822 CH471090 BC136618 BC171770 AB037843 AAQR03116405 AAQR03116406 AAQR03116407 NDHI03003456 PNJ45115.1 GCES01071539 JAR14784.1 KP869068 ALA62333.1 AADN04000037 AY093434 PNI68606.1 CABD030067347 CABD030067348 CABD030067349 PNI68608.1 PNJ45120.1 PNJ45122.1 GCES01071535 JAR14788.1
KQ971314 KYB29019.1 KYB29020.1 AAAB01008807 EDO64534.2 OUUW01000001 SPP74099.1 SPP74096.1 ABLF02039742 ABLF02039754 ABLF02047215 UFQT01000105 SSX20041.1 SSX20043.1 SSX20037.1 SSX20045.1 SSX20040.1 SSX20036.1 CVRI01000058 CRL02892.1 GEZM01007528 JAV95109.1 SSX20042.1 GEZM01007527 JAV95110.1 GGMR01009773 MBY22392.1 SSX20044.1 CRL02893.1 DS235862 EEB19234.1 SSX20038.1 GDIP01039686 JAM64029.1 LRGB01000327 KZS19731.1 GDIP01038538 JAM65177.1 GDIP01000470 JAN03246.1 GDIP01190159 JAJ33243.1 GDIP01082322 JAM21393.1 GDIP01035246 JAM68469.1 APGK01026655 APGK01026656 APGK01026657 APGK01026658 APGK01026659 KB740635 ENN79925.1 GDIP01152820 JAJ70582.1 GL732629 EFX69876.1 GDIP01082323 JAM21392.1 GDIP01202063 JAJ21339.1 KQ414736 KOC62087.1 GDIQ01001674 JAN93063.1 GDIP01038537 JAM65178.1 AJWK01012580 AJWK01012581 AJWK01012582 AJWK01012583 AJWK01012584 AJWK01012585 KB630556 ERL83673.1 KB632107 ERL88866.1 KU681442 AOR07222.1 JH818227 EKC24758.1 KQ423495 KOF72763.1 KOF72762.1 SSX20034.1 NIVC01002133 PAA60613.1 SSX20039.1 AGTP01090746 AGTP01090747 AGTP01090748 AACZ04057952 NBAG03000232 PNI68601.1 AK127272 AL158822 CH471090 BC136618 BC171770 AB037843 AAQR03116405 AAQR03116406 AAQR03116407 NDHI03003456 PNJ45115.1 GCES01071539 JAR14784.1 KP869068 ALA62333.1 AADN04000037 AY093434 PNI68606.1 CABD030067347 CABD030067348 CABD030067349 PNI68608.1 PNJ45120.1 PNJ45122.1 GCES01071535 JAR14788.1
Proteomes
UP000007151
UP000053240
UP000053268
UP000007266
UP000007062
UP000075880
+ More
UP000268350 UP000192223 UP000007819 UP000183832 UP000009046 UP000076858 UP000019118 UP000000305 UP000053825 UP000092461 UP000030742 UP000005408 UP000053454 UP000001038 UP000215902 UP000261420 UP000261400 UP000233040 UP000005215 UP000002277 UP000233020 UP000005640 UP000005225 UP000233160 UP000233180 UP000233200 UP000265000 UP000265140 UP000265180 UP000000539 UP000265200 UP000233120 UP000192224 UP000001519 UP000261540 UP000261440
UP000268350 UP000192223 UP000007819 UP000183832 UP000009046 UP000076858 UP000019118 UP000000305 UP000053825 UP000092461 UP000030742 UP000005408 UP000053454 UP000001038 UP000215902 UP000261420 UP000261400 UP000233040 UP000005215 UP000002277 UP000233020 UP000005640 UP000005225 UP000233160 UP000233180 UP000233200 UP000265000 UP000265140 UP000265180 UP000000539 UP000265200 UP000233120 UP000192224 UP000001519 UP000261540 UP000261440
Interpro
SUPFAM
SSF51735
SSF51735
ProteinModelPortal
A0A212EY19
A0A194QWH0
A0A194PY46
A0A139WLY1
A0A139WM47
A7UR92
+ More
A0A182IR49 A0A3B0JKV1 A0A1W4XHF4 A0A3B0JLH8 J9K6K2 A0A336LSZ5 A0A336LTP8 A0A336LUK8 A0A336LQC2 A0A336LQ39 A0A336LPP9 A0A1J1ITI9 A0A1Y1NBK9 A0A336LPT4 A0A1Y1NE12 A0A2S2NZ98 A0A336LPM5 A0A1J1IRM2 E0W0S8 A0A336M243 A0A0P5ZTV5 A0A162QJ56 A0A0P5ZX15 A0A0P6CR92 A0A0P5B3Q8 A0A0P5XMA2 A0A0P6A468 N6UDZ1 A0A0P5DU19 E9HEH7 A0A0P5WSA0 A0A0P5AC24 A0A0L7QTU4 A0A0P6IQA5 A0A0P6ATN0 A0A1B0CHQ2 U4TUR2 U4UE65 A0A1C9TA78 K1PSX5 A0A0L8G7B1 A0A0L8G6V3 A0A3B3I015 A0A336LPL1 A0A267EGF8 A0A3B4VM99 A0A3B4ZIJ9 A0A336LTT0 A0A2K5RPJ3 I3N3P6 A0A2I3S2J6 A0A2K5CTR7 Q5JUK3-3 H0WYR9 A0A2J8UIJ8 A0A2K6GK25 A0A2K6K4N9 A0A2K6NPW6 A0A146VBH1 A0A0K2GMW5 A0A3P8XE41 A0A3P9KR47 A0A3B4VM72 A0A3P8XB84 E1BW18 Q8QFV0 A0A3P9IIP2 A0A2K6DE59 A0A1W5ACB6 A0A3B4VIR6 A0A2K5RPF5 A0A2K5CTT5 A0A2I3T3B9 A0A3P8XB73 A0A2I2YBE5 A0A2K5RPK7 A0A287D7E5 A0A2J8NA34 A0A3B3RM10 A0A2K5CTS9 Q5JUK3-2 A0A2K6K4Q0 A0A0D9SFC8 A0A2I2YX38 G3QR87 A0A2J8UIP1 A0A2J8UIN6 A0A3B4EPN9 A0A2K5CTS6 A0A2K6GK47 A0A3B3RND9 A0A2K6K4M7 A0A2K6NPX6 A0A146VC13
A0A182IR49 A0A3B0JKV1 A0A1W4XHF4 A0A3B0JLH8 J9K6K2 A0A336LSZ5 A0A336LTP8 A0A336LUK8 A0A336LQC2 A0A336LQ39 A0A336LPP9 A0A1J1ITI9 A0A1Y1NBK9 A0A336LPT4 A0A1Y1NE12 A0A2S2NZ98 A0A336LPM5 A0A1J1IRM2 E0W0S8 A0A336M243 A0A0P5ZTV5 A0A162QJ56 A0A0P5ZX15 A0A0P6CR92 A0A0P5B3Q8 A0A0P5XMA2 A0A0P6A468 N6UDZ1 A0A0P5DU19 E9HEH7 A0A0P5WSA0 A0A0P5AC24 A0A0L7QTU4 A0A0P6IQA5 A0A0P6ATN0 A0A1B0CHQ2 U4TUR2 U4UE65 A0A1C9TA78 K1PSX5 A0A0L8G7B1 A0A0L8G6V3 A0A3B3I015 A0A336LPL1 A0A267EGF8 A0A3B4VM99 A0A3B4ZIJ9 A0A336LTT0 A0A2K5RPJ3 I3N3P6 A0A2I3S2J6 A0A2K5CTR7 Q5JUK3-3 H0WYR9 A0A2J8UIJ8 A0A2K6GK25 A0A2K6K4N9 A0A2K6NPW6 A0A146VBH1 A0A0K2GMW5 A0A3P8XE41 A0A3P9KR47 A0A3B4VM72 A0A3P8XB84 E1BW18 Q8QFV0 A0A3P9IIP2 A0A2K6DE59 A0A1W5ACB6 A0A3B4VIR6 A0A2K5RPF5 A0A2K5CTT5 A0A2I3T3B9 A0A3P8XB73 A0A2I2YBE5 A0A2K5RPK7 A0A287D7E5 A0A2J8NA34 A0A3B3RM10 A0A2K5CTS9 Q5JUK3-2 A0A2K6K4Q0 A0A0D9SFC8 A0A2I2YX38 G3QR87 A0A2J8UIP1 A0A2J8UIN6 A0A3B4EPN9 A0A2K5CTS6 A0A2K6GK47 A0A3B3RND9 A0A2K6K4M7 A0A2K6NPX6 A0A146VC13
PDB
5U76
E-value=0,
Score=3137
Ontologies
GO
Topology
Subcellular location
Cell membrane
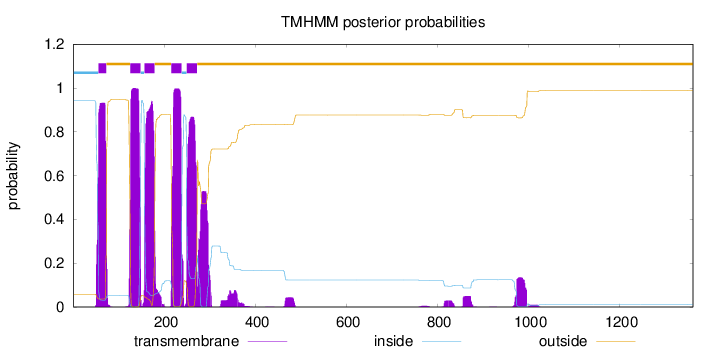
Length:
1362
Number of predicted TMHs:
5
Exp number of AAs in TMHs:
122.88911
Exp number, first 60 AAs:
6.52533
Total prob of N-in:
0.94245
inside
1 - 55
TMhelix
56 - 73
outside
74 - 125
TMhelix
126 - 148
inside
149 - 156
TMhelix
157 - 179
outside
180 - 215
TMhelix
216 - 238
inside
239 - 249
TMhelix
250 - 272
outside
273 - 1362
Population Genetic Test Statistics
Pi
18.460882
Theta
18.22277
Tajima's D
0.300062
CLR
0.087142
CSRT
0.464626768661567
Interpretation
Uncertain
