Gene
KWMTBOMO00346
Pre Gene Modal
BGIBMGA000523
Annotation
PREDICTED:_tyrosine-protein_kinase_receptor_TYRO3-like_[Papilio_polytes]
Location in the cell
Extracellular Reliability : 1.421 Mitochondrial Reliability : 1.507
Sequence
CDS
ATGTTCTCTAAAGCGAAATTTATCTTCAGTTTACTCTTACTGTCTTTGATTCTAATAAATCAAGCTGATGGTAGACGAGGGCGACCCAGGAGCAGGACCAAGTCGAAGGTGCAGATCGGGTTGCCGATCACGGGCAAGTACCGAGACCCAGAATCCGACCAATACTACAATAATAATGATGGAGCAAAGATTTTACTGGCCTCACACTTTGATCTTGAGTATTCACTGGGGCACAAGATCGCGTTCCTCTGCATCGCGAAGGGTAACCCGCGGCCGCATATCACGTGGTTCAAGGACGGAGTGGAAATATTCGCTCATCATTATTTACACATTCACGAATGGCCAATTGGCGAAGATCGTGTGAAATCTAAGATCGAGATCGATCCAGCTACACAGATGGACGCCGGCGTTTACGAGTGCACTGCCGACAACATGTATTCTATTGACCGTCGCTCCTTCAAGACTGACTTCTCTATCGCTTTTGACTGA
Protein
MFSKAKFIFSLLLLSLILINQADGRRGRPRSRTKSKVQIGLPITGKYRDPESDQYYNNNDGAKILLASHFDLEYSLGHKIAFLCIAKGNPRPHITWFKDGVEIFAHHYLHIHEWPIGEDRVKSKIEIDPATQMDAGVYECTADNMYSIDRRSFKTDFSIAFD
Summary
Similarity
Belongs to the EMP24/GP25L family.
Uniprot
H9ITE3
A0A2H1W1K6
A0A2A4IV12
A0A194PRY3
A0A194QVG5
A0A212F458
+ More
A0A2P8XNS6 A0A1J1IBG4 B4L190 A0A0L7RA19 A0A1B0CUR9 B4LC65 A0A2A3ED21 A0A087ZSH1 A0A3B0JUL3 W8B4E9 B4N5C7 Q2LZ42 B4GZM5 B4J2V8 A0A195D5S8 A0A0K8UEG4 Q8SX06 A0A1W4W6F8 A0A0A1WIF6 A0A158NHE8 A0A034WUG3 A0A154PD28 B4PF21 B4HEB7 B3NGT3 B4QPR7 B3M3Y3 A0A0M4EYS7 A0A1I8MCV4 A0A1B0G3J2 A0A1A9YEM3 A0A1A9VMY2 A0A1B0B199 A0A1B0A2H5 A0A182PGY3 A0A182JTM3 A0A1B6C4F5 A0A1Y9G869 U5EKX6 A0A182MR08 A0A2C9GQU2 A0A182U5X9 Q7PKQ6 A0A1I8Q863 A0A182YB79 B0WXT6 A0A2C9H8F6 A0A182MTG1 A0A182NQC9 A0A182T8U3 A0A1Q3FPG8 Q0IES1 A0A023EHQ1 A0A1Y1L5L6 A0A0M8ZT96 A0A3L8DH20 A0A026WTD6 A0A1B6IT32 A0A195E297 A0A195FWP1 A0A151XFD4 A0A1W4WHG6 A0A195B075 A0A1B6FE08 E0VRG0 A0A310SHY2 A0A0L0C0F0 A0A1B6LVT5 A0A1A9W4M1 A0A1B6KQZ2 A0A0A9YR49 A0A0A9YV97 W5JBX8 A0A067RGK6 N6UF32 A0A0C9RVI6 A0A2S2PI58 D6WLT8 J9JLV4 A0A2H8TZR9 A0A232FA72 K7JTG0 A0A0K8TL27 A0A1B0DAD4 R4G3T7 A0A336JXB0 A0A182R4Z4 A0A084W541 A0A2J7RR26 A0A336K0Q6 T1GH28 A0A1D2MLT4 A0A226EIX5
A0A2P8XNS6 A0A1J1IBG4 B4L190 A0A0L7RA19 A0A1B0CUR9 B4LC65 A0A2A3ED21 A0A087ZSH1 A0A3B0JUL3 W8B4E9 B4N5C7 Q2LZ42 B4GZM5 B4J2V8 A0A195D5S8 A0A0K8UEG4 Q8SX06 A0A1W4W6F8 A0A0A1WIF6 A0A158NHE8 A0A034WUG3 A0A154PD28 B4PF21 B4HEB7 B3NGT3 B4QPR7 B3M3Y3 A0A0M4EYS7 A0A1I8MCV4 A0A1B0G3J2 A0A1A9YEM3 A0A1A9VMY2 A0A1B0B199 A0A1B0A2H5 A0A182PGY3 A0A182JTM3 A0A1B6C4F5 A0A1Y9G869 U5EKX6 A0A182MR08 A0A2C9GQU2 A0A182U5X9 Q7PKQ6 A0A1I8Q863 A0A182YB79 B0WXT6 A0A2C9H8F6 A0A182MTG1 A0A182NQC9 A0A182T8U3 A0A1Q3FPG8 Q0IES1 A0A023EHQ1 A0A1Y1L5L6 A0A0M8ZT96 A0A3L8DH20 A0A026WTD6 A0A1B6IT32 A0A195E297 A0A195FWP1 A0A151XFD4 A0A1W4WHG6 A0A195B075 A0A1B6FE08 E0VRG0 A0A310SHY2 A0A0L0C0F0 A0A1B6LVT5 A0A1A9W4M1 A0A1B6KQZ2 A0A0A9YR49 A0A0A9YV97 W5JBX8 A0A067RGK6 N6UF32 A0A0C9RVI6 A0A2S2PI58 D6WLT8 J9JLV4 A0A2H8TZR9 A0A232FA72 K7JTG0 A0A0K8TL27 A0A1B0DAD4 R4G3T7 A0A336JXB0 A0A182R4Z4 A0A084W541 A0A2J7RR26 A0A336K0Q6 T1GH28 A0A1D2MLT4 A0A226EIX5
Pubmed
19121390
26354079
22118469
29403074
17994087
24495485
+ More
15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 21347285 25348373 17550304 22936249 25315136 12364791 14747013 17210077 25244985 17510324 24945155 26483478 28004739 30249741 24508170 20566863 26108605 25401762 26823975 20920257 23761445 24845553 23537049 18362917 19820115 28648823 20075255 26369729 24438588 27289101
15632085 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 26109357 26109356 25830018 21347285 25348373 17550304 22936249 25315136 12364791 14747013 17210077 25244985 17510324 24945155 26483478 28004739 30249741 24508170 20566863 26108605 25401762 26823975 20920257 23761445 24845553 23537049 18362917 19820115 28648823 20075255 26369729 24438588 27289101
EMBL
BABH01007118
ODYU01005652
SOQ46722.1
NWSH01006243
PCG63561.1
KQ459594
+ More
KPI96067.1 KQ461108 KPJ09314.1 AGBW02010446 OWR48493.1 PYGN01001632 PSN33661.1 CVRI01000047 CRK97621.1 CH933809 EDW19272.2 KQ414619 KOC67722.1 AJWK01029612 CH940647 EDW70893.1 KZ288280 PBC29645.1 OUUW01000002 SPP77046.1 GAMC01018394 JAB88161.1 CH964101 EDW79566.1 CH379069 EAL29667.2 CH479199 EDW29452.1 CH916366 EDV97128.1 KQ976870 KYN07789.1 GDHF01027346 JAI24968.1 AE014296 AY094920 KX532096 AAF50080.2 AAM11273.1 ANY27906.1 GBXI01015831 JAC98460.1 ADTU01015730 GAKP01001117 GAKP01001116 JAC57835.1 KQ434868 KZC09168.1 CM000159 EDW94103.1 CH480815 EDW41071.1 CH954178 EDV51390.1 CM000363 CM002912 EDX10058.1 KMY98955.1 CH902618 EDV39317.1 CP012525 ALC43712.1 CCAG010011629 JXJN01007030 JXJN01007031 GEDC01029014 GEDC01021073 JAS08284.1 JAS16225.1 GANO01005050 JAB54821.1 AXCM01000787 APCN01000879 AAAB01008987 EAA43173.4 DS232173 EDS36746.1 AXCM01007623 GFDL01005546 JAV29499.1 CH477486 EAT40133.1 JXUM01110351 GAPW01005219 KQ565404 JAC08379.1 KXJ71019.1 GEZM01068636 JAV66846.1 KQ435903 KOX69079.1 QOIP01000008 RLU19740.1 KK107109 EZA59213.1 GECU01017676 JAS90030.1 KQ979763 KYN19201.1 KQ981208 KYN44846.1 KQ982194 KYQ59102.1 KQ976694 KYM77609.1 GECZ01021556 JAS48213.1 DS235465 EEB15966.1 KQ760974 OAD58571.1 JRES01001072 KNC25773.1 GEBQ01012188 JAT27789.1 GEBQ01026114 JAT13863.1 GBHO01008072 JAG35532.1 GBHO01008073 GBRD01002174 GDHC01018710 JAG35531.1 JAG63647.1 JAP99918.1 ADMH02001952 ETN60309.1 KK852521 KDR22123.1 APGK01037211 KB740948 KB632348 ENN77267.1 ERL93181.1 GBYB01012945 JAG82712.1 GGMR01016455 MBY29074.1 KQ971343 EFA04163.1 ABLF02032167 GFXV01007959 MBW19764.1 NNAY01000548 OXU27746.1 GDAI01003008 JAI14595.1 AJVK01013275 ACPB03002429 GAHY01001935 JAA75575.1 UFQS01000016 UFQT01000016 SSW97426.1 SSX17812.1 ATLV01020477 KE525302 KFB45335.1 NEVH01000611 PNF43289.1 UFQS01000038 UFQT01000038 SSW98031.1 SSX18417.1 CAQQ02390954 LJIJ01000902 ODM93851.1 LNIX01000003 OXA56711.1
KPI96067.1 KQ461108 KPJ09314.1 AGBW02010446 OWR48493.1 PYGN01001632 PSN33661.1 CVRI01000047 CRK97621.1 CH933809 EDW19272.2 KQ414619 KOC67722.1 AJWK01029612 CH940647 EDW70893.1 KZ288280 PBC29645.1 OUUW01000002 SPP77046.1 GAMC01018394 JAB88161.1 CH964101 EDW79566.1 CH379069 EAL29667.2 CH479199 EDW29452.1 CH916366 EDV97128.1 KQ976870 KYN07789.1 GDHF01027346 JAI24968.1 AE014296 AY094920 KX532096 AAF50080.2 AAM11273.1 ANY27906.1 GBXI01015831 JAC98460.1 ADTU01015730 GAKP01001117 GAKP01001116 JAC57835.1 KQ434868 KZC09168.1 CM000159 EDW94103.1 CH480815 EDW41071.1 CH954178 EDV51390.1 CM000363 CM002912 EDX10058.1 KMY98955.1 CH902618 EDV39317.1 CP012525 ALC43712.1 CCAG010011629 JXJN01007030 JXJN01007031 GEDC01029014 GEDC01021073 JAS08284.1 JAS16225.1 GANO01005050 JAB54821.1 AXCM01000787 APCN01000879 AAAB01008987 EAA43173.4 DS232173 EDS36746.1 AXCM01007623 GFDL01005546 JAV29499.1 CH477486 EAT40133.1 JXUM01110351 GAPW01005219 KQ565404 JAC08379.1 KXJ71019.1 GEZM01068636 JAV66846.1 KQ435903 KOX69079.1 QOIP01000008 RLU19740.1 KK107109 EZA59213.1 GECU01017676 JAS90030.1 KQ979763 KYN19201.1 KQ981208 KYN44846.1 KQ982194 KYQ59102.1 KQ976694 KYM77609.1 GECZ01021556 JAS48213.1 DS235465 EEB15966.1 KQ760974 OAD58571.1 JRES01001072 KNC25773.1 GEBQ01012188 JAT27789.1 GEBQ01026114 JAT13863.1 GBHO01008072 JAG35532.1 GBHO01008073 GBRD01002174 GDHC01018710 JAG35531.1 JAG63647.1 JAP99918.1 ADMH02001952 ETN60309.1 KK852521 KDR22123.1 APGK01037211 KB740948 KB632348 ENN77267.1 ERL93181.1 GBYB01012945 JAG82712.1 GGMR01016455 MBY29074.1 KQ971343 EFA04163.1 ABLF02032167 GFXV01007959 MBW19764.1 NNAY01000548 OXU27746.1 GDAI01003008 JAI14595.1 AJVK01013275 ACPB03002429 GAHY01001935 JAA75575.1 UFQS01000016 UFQT01000016 SSW97426.1 SSX17812.1 ATLV01020477 KE525302 KFB45335.1 NEVH01000611 PNF43289.1 UFQS01000038 UFQT01000038 SSW98031.1 SSX18417.1 CAQQ02390954 LJIJ01000902 ODM93851.1 LNIX01000003 OXA56711.1
Proteomes
UP000005204
UP000218220
UP000053268
UP000053240
UP000007151
UP000245037
+ More
UP000183832 UP000009192 UP000053825 UP000092461 UP000008792 UP000242457 UP000005203 UP000268350 UP000007798 UP000001819 UP000008744 UP000001070 UP000078542 UP000000803 UP000192221 UP000005205 UP000076502 UP000002282 UP000001292 UP000008711 UP000000304 UP000007801 UP000092553 UP000095301 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000075885 UP000075881 UP000069272 UP000075883 UP000075840 UP000075902 UP000007062 UP000095300 UP000076408 UP000002320 UP000076407 UP000075884 UP000075901 UP000008820 UP000069940 UP000249989 UP000053105 UP000279307 UP000053097 UP000078492 UP000078541 UP000075809 UP000192223 UP000078540 UP000009046 UP000037069 UP000091820 UP000000673 UP000027135 UP000019118 UP000030742 UP000007266 UP000007819 UP000215335 UP000002358 UP000092462 UP000015103 UP000075900 UP000030765 UP000235965 UP000015102 UP000094527 UP000198287
UP000183832 UP000009192 UP000053825 UP000092461 UP000008792 UP000242457 UP000005203 UP000268350 UP000007798 UP000001819 UP000008744 UP000001070 UP000078542 UP000000803 UP000192221 UP000005205 UP000076502 UP000002282 UP000001292 UP000008711 UP000000304 UP000007801 UP000092553 UP000095301 UP000092444 UP000092443 UP000078200 UP000092460 UP000092445 UP000075885 UP000075881 UP000069272 UP000075883 UP000075840 UP000075902 UP000007062 UP000095300 UP000076408 UP000002320 UP000076407 UP000075884 UP000075901 UP000008820 UP000069940 UP000249989 UP000053105 UP000279307 UP000053097 UP000078492 UP000078541 UP000075809 UP000192223 UP000078540 UP000009046 UP000037069 UP000091820 UP000000673 UP000027135 UP000019118 UP000030742 UP000007266 UP000007819 UP000215335 UP000002358 UP000092462 UP000015103 UP000075900 UP000030765 UP000235965 UP000015102 UP000094527 UP000198287
Interpro
Gene 3D
ProteinModelPortal
H9ITE3
A0A2H1W1K6
A0A2A4IV12
A0A194PRY3
A0A194QVG5
A0A212F458
+ More
A0A2P8XNS6 A0A1J1IBG4 B4L190 A0A0L7RA19 A0A1B0CUR9 B4LC65 A0A2A3ED21 A0A087ZSH1 A0A3B0JUL3 W8B4E9 B4N5C7 Q2LZ42 B4GZM5 B4J2V8 A0A195D5S8 A0A0K8UEG4 Q8SX06 A0A1W4W6F8 A0A0A1WIF6 A0A158NHE8 A0A034WUG3 A0A154PD28 B4PF21 B4HEB7 B3NGT3 B4QPR7 B3M3Y3 A0A0M4EYS7 A0A1I8MCV4 A0A1B0G3J2 A0A1A9YEM3 A0A1A9VMY2 A0A1B0B199 A0A1B0A2H5 A0A182PGY3 A0A182JTM3 A0A1B6C4F5 A0A1Y9G869 U5EKX6 A0A182MR08 A0A2C9GQU2 A0A182U5X9 Q7PKQ6 A0A1I8Q863 A0A182YB79 B0WXT6 A0A2C9H8F6 A0A182MTG1 A0A182NQC9 A0A182T8U3 A0A1Q3FPG8 Q0IES1 A0A023EHQ1 A0A1Y1L5L6 A0A0M8ZT96 A0A3L8DH20 A0A026WTD6 A0A1B6IT32 A0A195E297 A0A195FWP1 A0A151XFD4 A0A1W4WHG6 A0A195B075 A0A1B6FE08 E0VRG0 A0A310SHY2 A0A0L0C0F0 A0A1B6LVT5 A0A1A9W4M1 A0A1B6KQZ2 A0A0A9YR49 A0A0A9YV97 W5JBX8 A0A067RGK6 N6UF32 A0A0C9RVI6 A0A2S2PI58 D6WLT8 J9JLV4 A0A2H8TZR9 A0A232FA72 K7JTG0 A0A0K8TL27 A0A1B0DAD4 R4G3T7 A0A336JXB0 A0A182R4Z4 A0A084W541 A0A2J7RR26 A0A336K0Q6 T1GH28 A0A1D2MLT4 A0A226EIX5
A0A2P8XNS6 A0A1J1IBG4 B4L190 A0A0L7RA19 A0A1B0CUR9 B4LC65 A0A2A3ED21 A0A087ZSH1 A0A3B0JUL3 W8B4E9 B4N5C7 Q2LZ42 B4GZM5 B4J2V8 A0A195D5S8 A0A0K8UEG4 Q8SX06 A0A1W4W6F8 A0A0A1WIF6 A0A158NHE8 A0A034WUG3 A0A154PD28 B4PF21 B4HEB7 B3NGT3 B4QPR7 B3M3Y3 A0A0M4EYS7 A0A1I8MCV4 A0A1B0G3J2 A0A1A9YEM3 A0A1A9VMY2 A0A1B0B199 A0A1B0A2H5 A0A182PGY3 A0A182JTM3 A0A1B6C4F5 A0A1Y9G869 U5EKX6 A0A182MR08 A0A2C9GQU2 A0A182U5X9 Q7PKQ6 A0A1I8Q863 A0A182YB79 B0WXT6 A0A2C9H8F6 A0A182MTG1 A0A182NQC9 A0A182T8U3 A0A1Q3FPG8 Q0IES1 A0A023EHQ1 A0A1Y1L5L6 A0A0M8ZT96 A0A3L8DH20 A0A026WTD6 A0A1B6IT32 A0A195E297 A0A195FWP1 A0A151XFD4 A0A1W4WHG6 A0A195B075 A0A1B6FE08 E0VRG0 A0A310SHY2 A0A0L0C0F0 A0A1B6LVT5 A0A1A9W4M1 A0A1B6KQZ2 A0A0A9YR49 A0A0A9YV97 W5JBX8 A0A067RGK6 N6UF32 A0A0C9RVI6 A0A2S2PI58 D6WLT8 J9JLV4 A0A2H8TZR9 A0A232FA72 K7JTG0 A0A0K8TL27 A0A1B0DAD4 R4G3T7 A0A336JXB0 A0A182R4Z4 A0A084W541 A0A2J7RR26 A0A336K0Q6 T1GH28 A0A1D2MLT4 A0A226EIX5
PDB
5XWU
E-value=8.42264e-06,
Score=112
Ontologies
GO
PANTHER
Topology
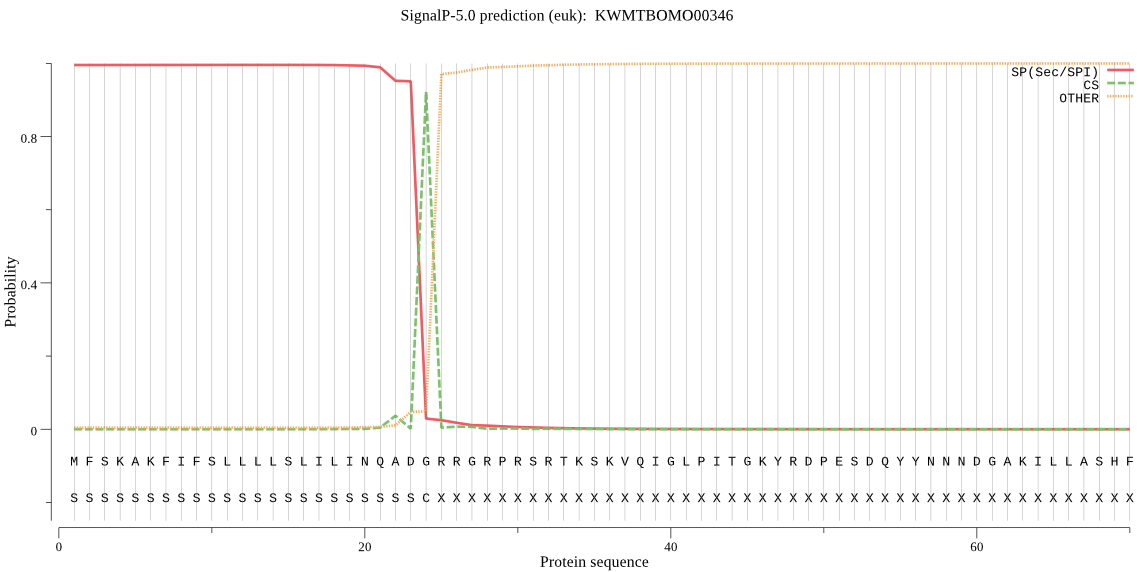
SignalP
Position: 1 - 24,
Likelihood: 0.995515
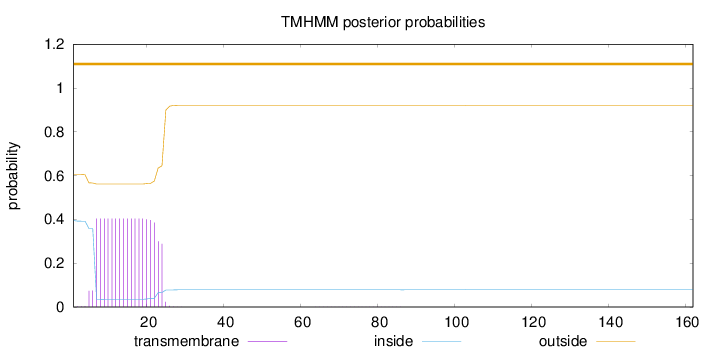
Length:
162
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
7.21023
Exp number, first 60 AAs:
7.20623
Total prob of N-in:
0.39499
outside
1 - 162
Population Genetic Test Statistics
Pi
6.908778
Theta
6.059014
Tajima's D
0.568513
CLR
0.024413
CSRT
0.56037198140093
Interpretation
Uncertain
