Gene
KWMTBOMO00340 Validated by peptides from experiments
Pre Gene Modal
BGIBMGA000701
Annotation
catalase_[Bombyx_mori]
Full name
Catalase
Location in the cell
Peroxisomal Reliability : 3.293
Sequence
CDS
ATGGCTTCAAGAGACCCAGCTACTGACCAACTCATTAACTACAAGAAAACTTTAAAGGATTCTCCTGGTTTCATCACCACTAAATCTGGAGCTCCTGTTGGAATCAAAACGGCGATACAAACGGTGGGCAAGAATGGTCCAGCCTTATTACAGGATGTTAACTTTCTTGATGAAATGTCATCCTTTGACAGGGAGCGTATTCCAGAACGAGTGGTACATGCTAAAGGAGCTGGAGCTTTTGGGTACTTTGAAGTAACTCATGACATCACCAAGTACAGTGCTGCCAAAGTATTTGAGTCCATAGGCAAAAGGACACCGATTGCTGTTAGATTCTCAACAGTTGGTGGAGAGAGTGGATCTGCTGATACTGTTCGTGACCCTCGAGGTTTTGCTGTAAAGTTCTATACTGATGATGGAGTGTGGGATTTAGTTGGAAATAATACTCCCATTTTCTTTATAAGAGATCCAACATTATTCCCGAGTTTTATCCATACTCAGAAGAGAAACCCTGCAACACATCTGAAAGATCCAGACATGTTTTGGGACTTTTTGACCTTAAGACCAGAGACCATCCATCAACTTCTTTACTTGTTTGGAGACCGTGGTATTCCTGATGGCTACAGACATATGAATGGATATGGTTCCCATACATTCAAGCTTGTGAATTCTCAAGGTGTTGGGTACTGGGTAAAATTCCACTATAAGACTAACCAGGGGATTAAAAACTTATCAGTTGACAAAGCAGGTGAACTGGCATCAACTGATCCTGATTATTCCATTAGAGATCTGTATAATTCCATTGCAAAGGGTGATTACCCATCATGGACTTTTTACATTCAGGTGATGACTATGGCTCAAGCAGAAAGCTGTAAATTTAATCCGTTTGACCTAACTAAGATCTGGCCACATGCAGAATATCCTTTAATTCCTGTTGGTAAATTGGTTTTGGATAGAAACCCTAAGAATTACTTTGCTGAAGTTGAACAAATTGCTTTCAGTCCATCTAATTTAGTACCAGGAATTGAGCCATCTCCAGACAAAATGTTACAAGGAAGACTGTTTGCCTATAGCGATACACATCGCCATAGACTTGGTGCTAACTACCTCCAAATTCCTGTAAATTGTCCATACAAAGTTGCTGTTTCCACCTACCAGCGTGATGGACCTCAAGCTATTCACAATCAAGACGACTGCCCTAACTATTTCCCCAACTCTTTTTCTGGACCACAAGAATGTCCACGTGCTCAGCGTTTGCAACCTCGATACAATGTCGGCGGAGATGTAGACCGGTATGATAGTGGCCAGACTGAGGACAACTTCTCCCAAGCGACAGCACTGTATAAACAAGTTTTTGATGATGCAGCTAAACAACGAGCTATTGCCAATATAGTAGATCACCTCAAGGATGCTGCTGCGTTTATTCAAGAAAGAGCTATCAAGATCTTCTCTCAGGTACATCCTGAACTTGGAAATAAAGTAGCAGCTGGTTTAGCTCCTTACAAAAAATATCATGCAAATTTGTAA
Protein
MASRDPATDQLINYKKTLKDSPGFITTKSGAPVGIKTAIQTVGKNGPALLQDVNFLDEMSSFDRERIPERVVHAKGAGAFGYFEVTHDITKYSAAKVFESIGKRTPIAVRFSTVGGESGSADTVRDPRGFAVKFYTDDGVWDLVGNNTPIFFIRDPTLFPSFIHTQKRNPATHLKDPDMFWDFLTLRPETIHQLLYLFGDRGIPDGYRHMNGYGSHTFKLVNSQGVGYWVKFHYKTNQGIKNLSVDKAGELASTDPDYSIRDLYNSIAKGDYPSWTFYIQVMTMAQAESCKFNPFDLTKIWPHAEYPLIPVGKLVLDRNPKNYFAEVEQIAFSPSNLVPGIEPSPDKMLQGRLFAYSDTHRHRLGANYLQIPVNCPYKVAVSTYQRDGPQAIHNQDDCPNYFPNSFSGPQECPRAQRLQPRYNVGGDVDRYDSGQTEDNFSQATALYKQVFDDAAKQRAIANIVDHLKDAAAFIQERAIKIFSQVHPELGNKVAAGLAPYKKYHANL
Summary
Description
Occurs in almost all aerobically respiring organisms and serves to protect cells from the toxic effects of hydrogen peroxide.
Catalytic Activity
2 H2O2 = 2 H2O + O2
Cofactor
heme
Subunit
Homotetramer.
Similarity
Belongs to the catalase family.
Keywords
Complete proteome
Direct protein sequencing
Heme
Hydrogen peroxide
Iron
Metal-binding
Oxidoreductase
Peroxidase
Peroxisome
Reference proteome
Feature
chain Catalase
Uniprot
H9ITX1
Q68AP5
A0A194QVH0
A0A1V0QH47
A0A194PRI4
A0A2R2PTF8
+ More
H9BEW3 G5CDD8 I4DMH7 A0A2H1WQ68 A0A286RZM9 A0A2Z4N440 H9U5T8 S4PA78 A0A212F436 A0A0L7KXM9 A0A2S0RQP9 A0A0L7KZY0 F4MI31 A0A1B0CRV7 A0A1I8PLT9 A8CWF1 A0A3B0JYK5 A0A0L0C8Y9 A0A0K8TMT3 A0A1I8MF15 B4PIY6 A0A1I8PLM9 B4QPW5 T1PCG9 B4IFV9 B3NHN7 A0A1L8EEQ5 B4KXS2 Q2M0D6 A0A0N9E6Y9 A0A1L8EEC3 A0A034WTI4 A0A0C9Q530 B4N4F5 A0A0M4EK05 B3M869 A0A336N5N6 A0A0A1WYM5 P17336 A0A1W4UPD3 B4J1T4 W8C3S7 A0A0K8VAH0 T1E775 J9HGP0 A0A1L7LQD2 A0A182FDJ3 A0A023EUF0 Q16J86 A0A2M4BKY1 A0A1L8E475 A0A182GFN9 G9BQS7 A0A087ZC72 A0A2M4A193 A0A084WKX1 A0A1L8E4H4 K4N1L2 B4LFX2 A0A182IJJ1 E0YL15 A0A2M3YYC6 A0A2M3ZFP9 A0A2M4A1E0 A0A2M4CPT1 A0A2J7REJ5 A0A2M3YYJ7 A0A182WS20 A3E807 A0A182HQG6 A0A182UXY3 A0A1J1I836 A0A2M4CV05 A3E7Z8 A0A2M4CTV7 A0A182LDW5 A0A182PEB9 A3E800 A0A182QZS4 E2AJX6 A0A1B3PEJ4 A0A2M4A121 W5JER1 A0A067RLP1 A0A2M4A0F7 A0A1Q3FPN2 A0A131XJ28 G9C5E5 A0A224YPR9 A0A182N228 A0A224YJA5 A0A224YJ98 L7M7X5 A0A2U8T6B2
H9BEW3 G5CDD8 I4DMH7 A0A2H1WQ68 A0A286RZM9 A0A2Z4N440 H9U5T8 S4PA78 A0A212F436 A0A0L7KXM9 A0A2S0RQP9 A0A0L7KZY0 F4MI31 A0A1B0CRV7 A0A1I8PLT9 A8CWF1 A0A3B0JYK5 A0A0L0C8Y9 A0A0K8TMT3 A0A1I8MF15 B4PIY6 A0A1I8PLM9 B4QPW5 T1PCG9 B4IFV9 B3NHN7 A0A1L8EEQ5 B4KXS2 Q2M0D6 A0A0N9E6Y9 A0A1L8EEC3 A0A034WTI4 A0A0C9Q530 B4N4F5 A0A0M4EK05 B3M869 A0A336N5N6 A0A0A1WYM5 P17336 A0A1W4UPD3 B4J1T4 W8C3S7 A0A0K8VAH0 T1E775 J9HGP0 A0A1L7LQD2 A0A182FDJ3 A0A023EUF0 Q16J86 A0A2M4BKY1 A0A1L8E475 A0A182GFN9 G9BQS7 A0A087ZC72 A0A2M4A193 A0A084WKX1 A0A1L8E4H4 K4N1L2 B4LFX2 A0A182IJJ1 E0YL15 A0A2M3YYC6 A0A2M3ZFP9 A0A2M4A1E0 A0A2M4CPT1 A0A2J7REJ5 A0A2M3YYJ7 A0A182WS20 A3E807 A0A182HQG6 A0A182UXY3 A0A1J1I836 A0A2M4CV05 A3E7Z8 A0A2M4CTV7 A0A182LDW5 A0A182PEB9 A3E800 A0A182QZS4 E2AJX6 A0A1B3PEJ4 A0A2M4A121 W5JER1 A0A067RLP1 A0A2M4A0F7 A0A1Q3FPN2 A0A131XJ28 G9C5E5 A0A224YPR9 A0A182N228 A0A224YJA5 A0A224YJ98 L7M7X5 A0A2U8T6B2
EC Number
1.11.1.6
Pubmed
19121390
15763464
26354079
22651552
23622113
22118469
+ More
26227816 21569254 18194529 26108605 26369729 25315136 17994087 17550304 22936249 15632085 25348373 25830018 8660653 2362827 10731132 12537572 12537569 8500545 24495485 17510324 26760975 28076409 24945155 26483478 23441231 20920257 24438588 21103355 17284604 12364791 14747013 17210077 20966253 20798317 27521923 24845553 28049606 21835214 28797301 25576852
26227816 21569254 18194529 26108605 26369729 25315136 17994087 17550304 22936249 15632085 25348373 25830018 8660653 2362827 10731132 12537572 12537569 8500545 24495485 17510324 26760975 28076409 24945155 26483478 23441231 20920257 24438588 21103355 17284604 12364791 14747013 17210077 20966253 20798317 27521923 24845553 28049606 21835214 28797301 25576852
EMBL
BABH01007112
AB164195
BAD38853.1
KQ461108
KPJ09319.1
KY701971
+ More
ARE67828.1 KQ459594 KPI96061.1 KX079633 KX065288 ANA84065.1 JQ009332 AFC98367.1 JN051294 AEP40969.1 AK402495 BAM19117.1 ODYU01010180 SOQ55126.1 MF737386 ASX95436.1 MG397135 AWX63639.1 JQ663444 AFG31725.1 GAIX01003369 JAA89191.1 AGBW02010446 OWR48497.1 JTDY01004753 KOB67801.1 MF979105 AWA45968.1 JTDY01004115 KOB68584.1 EZ617712 ADH94605.1 AJWK01025349 AJWK01025350 AJWK01025351 AJWK01025352 EU124624 ABV60342.1 OUUW01000002 SPP76188.1 JRES01000836 KNC27864.1 GDAI01002147 JAI15456.1 CM000159 EDW94577.1 CM000363 CM002912 EDX10993.1 KMZ00447.1 KA645638 AFP60267.1 CH480834 EDW46546.1 CH954178 EDV51832.1 GFDG01001766 JAV17033.1 CH933809 EDW19779.1 CH379069 EAL30998.1 KR072552 ALF39394.1 GFDG01001805 JAV16994.1 GAKP01001864 JAC57088.1 GBYB01009173 JAG78940.1 CH964095 EDW79029.1 CP012525 ALC44426.1 CH902618 EDV41008.1 UFQT01002634 SSX33828.1 GBXI01010123 JAD04169.1 U00145 X52286 AE014296 AY084154 CH916366 EDV98014.1 GAMC01002637 JAC03919.1 GDHF01016433 JAI35881.1 GAMD01003292 JAA98298.1 CH478024 EJY58037.1 FX983163 BAV93783.1 GAPW01001047 JAC12551.1 EAT34333.1 GGFJ01004541 MBW53682.1 GFDF01000551 JAV13533.1 JXUM01010672 JXUM01010673 KQ560314 KXJ83094.1 HQ659100 AET14833.1 GGFK01001087 MBW34408.1 ATLV01024156 KE525350 KFB50865.1 GFDF01000550 JAV13534.1 JX513905 AFV36369.1 CH940647 EDW70371.1 HM062769 ADM26625.1 GGFM01000531 MBW21282.1 GGFM01006499 MBW27250.1 GGFK01001248 MBW34569.1 GGFL01003168 MBW67346.1 NEVH01004961 PNF39255.1 GGFM01000570 MBW21321.1 DQ980208 DQ980209 ABL09376.1 APCN01004603 CVRI01000043 CRK95906.1 GGFL01004500 MBW68678.1 DQ980199 DQ980200 DQ980204 DQ980205 DQ980206 DQ980207 AAAB01008948 ABL09367.1 ABL09372.1 EAA10427.4 GGFL01004531 MBW68709.1 DQ986315 DQ980201 DQ980202 DQ980203 DQ980210 ABK59975.1 ABL09369.1 AXCN02002351 GL440100 EFN66292.1 KT716445 AOG21082.1 GGFK01001195 MBW34516.1 ADMH02001698 ETN61375.1 KK852549 KDR21530.1 GGFK01000918 MBW34239.1 GFDL01005612 JAV29433.1 GEFH01002463 JAP66118.1 HQ851386 AEV89764.1 GFPF01006583 MAA17729.1 GFPF01006581 MAA17727.1 GFPF01006582 MAA17728.1 GACK01004914 JAA60120.1 KY953208 AWM72026.1
ARE67828.1 KQ459594 KPI96061.1 KX079633 KX065288 ANA84065.1 JQ009332 AFC98367.1 JN051294 AEP40969.1 AK402495 BAM19117.1 ODYU01010180 SOQ55126.1 MF737386 ASX95436.1 MG397135 AWX63639.1 JQ663444 AFG31725.1 GAIX01003369 JAA89191.1 AGBW02010446 OWR48497.1 JTDY01004753 KOB67801.1 MF979105 AWA45968.1 JTDY01004115 KOB68584.1 EZ617712 ADH94605.1 AJWK01025349 AJWK01025350 AJWK01025351 AJWK01025352 EU124624 ABV60342.1 OUUW01000002 SPP76188.1 JRES01000836 KNC27864.1 GDAI01002147 JAI15456.1 CM000159 EDW94577.1 CM000363 CM002912 EDX10993.1 KMZ00447.1 KA645638 AFP60267.1 CH480834 EDW46546.1 CH954178 EDV51832.1 GFDG01001766 JAV17033.1 CH933809 EDW19779.1 CH379069 EAL30998.1 KR072552 ALF39394.1 GFDG01001805 JAV16994.1 GAKP01001864 JAC57088.1 GBYB01009173 JAG78940.1 CH964095 EDW79029.1 CP012525 ALC44426.1 CH902618 EDV41008.1 UFQT01002634 SSX33828.1 GBXI01010123 JAD04169.1 U00145 X52286 AE014296 AY084154 CH916366 EDV98014.1 GAMC01002637 JAC03919.1 GDHF01016433 JAI35881.1 GAMD01003292 JAA98298.1 CH478024 EJY58037.1 FX983163 BAV93783.1 GAPW01001047 JAC12551.1 EAT34333.1 GGFJ01004541 MBW53682.1 GFDF01000551 JAV13533.1 JXUM01010672 JXUM01010673 KQ560314 KXJ83094.1 HQ659100 AET14833.1 GGFK01001087 MBW34408.1 ATLV01024156 KE525350 KFB50865.1 GFDF01000550 JAV13534.1 JX513905 AFV36369.1 CH940647 EDW70371.1 HM062769 ADM26625.1 GGFM01000531 MBW21282.1 GGFM01006499 MBW27250.1 GGFK01001248 MBW34569.1 GGFL01003168 MBW67346.1 NEVH01004961 PNF39255.1 GGFM01000570 MBW21321.1 DQ980208 DQ980209 ABL09376.1 APCN01004603 CVRI01000043 CRK95906.1 GGFL01004500 MBW68678.1 DQ980199 DQ980200 DQ980204 DQ980205 DQ980206 DQ980207 AAAB01008948 ABL09367.1 ABL09372.1 EAA10427.4 GGFL01004531 MBW68709.1 DQ986315 DQ980201 DQ980202 DQ980203 DQ980210 ABK59975.1 ABL09369.1 AXCN02002351 GL440100 EFN66292.1 KT716445 AOG21082.1 GGFK01001195 MBW34516.1 ADMH02001698 ETN61375.1 KK852549 KDR21530.1 GGFK01000918 MBW34239.1 GFDL01005612 JAV29433.1 GEFH01002463 JAP66118.1 HQ851386 AEV89764.1 GFPF01006583 MAA17729.1 GFPF01006581 MAA17727.1 GFPF01006582 MAA17728.1 GACK01004914 JAA60120.1 KY953208 AWM72026.1
Proteomes
UP000005204
UP000053240
UP000053268
UP000007151
UP000037510
UP000092461
+ More
UP000095300 UP000268350 UP000037069 UP000095301 UP000002282 UP000000304 UP000001292 UP000008711 UP000009192 UP000001819 UP000007798 UP000092553 UP000007801 UP000000803 UP000192221 UP000001070 UP000008820 UP000069272 UP000069940 UP000249989 UP000030765 UP000008792 UP000075880 UP000235965 UP000076407 UP000075840 UP000075903 UP000183832 UP000007062 UP000075882 UP000075885 UP000075886 UP000000311 UP000000673 UP000027135 UP000075884
UP000095300 UP000268350 UP000037069 UP000095301 UP000002282 UP000000304 UP000001292 UP000008711 UP000009192 UP000001819 UP000007798 UP000092553 UP000007801 UP000000803 UP000192221 UP000001070 UP000008820 UP000069272 UP000069940 UP000249989 UP000030765 UP000008792 UP000075880 UP000235965 UP000076407 UP000075840 UP000075903 UP000183832 UP000007062 UP000075882 UP000075885 UP000075886 UP000000311 UP000000673 UP000027135 UP000075884
Interpro
IPR020835
Catalase_sf
+ More
IPR011614 Catalase_core
IPR024711 Catalase_clade1/3
IPR002226 Catalase_haem_BS
IPR024708 Catalase_AS
IPR018028 Catalase
IPR010582 Catalase_immune_responsive
IPR040333 Catalase_3
IPR037060 Catalase_core_sf
IPR019786 Zinc_finger_PHD-type_CS
IPR019787 Znf_PHD-finger
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR011011 Znf_FYVE_PHD
IPR010916 TonB_box_CS
IPR011614 Catalase_core
IPR024711 Catalase_clade1/3
IPR002226 Catalase_haem_BS
IPR024708 Catalase_AS
IPR018028 Catalase
IPR010582 Catalase_immune_responsive
IPR040333 Catalase_3
IPR037060 Catalase_core_sf
IPR019786 Zinc_finger_PHD-type_CS
IPR019787 Znf_PHD-finger
IPR013083 Znf_RING/FYVE/PHD
IPR001965 Znf_PHD
IPR011011 Znf_FYVE_PHD
IPR010916 TonB_box_CS
Gene 3D
ProteinModelPortal
H9ITX1
Q68AP5
A0A194QVH0
A0A1V0QH47
A0A194PRI4
A0A2R2PTF8
+ More
H9BEW3 G5CDD8 I4DMH7 A0A2H1WQ68 A0A286RZM9 A0A2Z4N440 H9U5T8 S4PA78 A0A212F436 A0A0L7KXM9 A0A2S0RQP9 A0A0L7KZY0 F4MI31 A0A1B0CRV7 A0A1I8PLT9 A8CWF1 A0A3B0JYK5 A0A0L0C8Y9 A0A0K8TMT3 A0A1I8MF15 B4PIY6 A0A1I8PLM9 B4QPW5 T1PCG9 B4IFV9 B3NHN7 A0A1L8EEQ5 B4KXS2 Q2M0D6 A0A0N9E6Y9 A0A1L8EEC3 A0A034WTI4 A0A0C9Q530 B4N4F5 A0A0M4EK05 B3M869 A0A336N5N6 A0A0A1WYM5 P17336 A0A1W4UPD3 B4J1T4 W8C3S7 A0A0K8VAH0 T1E775 J9HGP0 A0A1L7LQD2 A0A182FDJ3 A0A023EUF0 Q16J86 A0A2M4BKY1 A0A1L8E475 A0A182GFN9 G9BQS7 A0A087ZC72 A0A2M4A193 A0A084WKX1 A0A1L8E4H4 K4N1L2 B4LFX2 A0A182IJJ1 E0YL15 A0A2M3YYC6 A0A2M3ZFP9 A0A2M4A1E0 A0A2M4CPT1 A0A2J7REJ5 A0A2M3YYJ7 A0A182WS20 A3E807 A0A182HQG6 A0A182UXY3 A0A1J1I836 A0A2M4CV05 A3E7Z8 A0A2M4CTV7 A0A182LDW5 A0A182PEB9 A3E800 A0A182QZS4 E2AJX6 A0A1B3PEJ4 A0A2M4A121 W5JER1 A0A067RLP1 A0A2M4A0F7 A0A1Q3FPN2 A0A131XJ28 G9C5E5 A0A224YPR9 A0A182N228 A0A224YJA5 A0A224YJ98 L7M7X5 A0A2U8T6B2
H9BEW3 G5CDD8 I4DMH7 A0A2H1WQ68 A0A286RZM9 A0A2Z4N440 H9U5T8 S4PA78 A0A212F436 A0A0L7KXM9 A0A2S0RQP9 A0A0L7KZY0 F4MI31 A0A1B0CRV7 A0A1I8PLT9 A8CWF1 A0A3B0JYK5 A0A0L0C8Y9 A0A0K8TMT3 A0A1I8MF15 B4PIY6 A0A1I8PLM9 B4QPW5 T1PCG9 B4IFV9 B3NHN7 A0A1L8EEQ5 B4KXS2 Q2M0D6 A0A0N9E6Y9 A0A1L8EEC3 A0A034WTI4 A0A0C9Q530 B4N4F5 A0A0M4EK05 B3M869 A0A336N5N6 A0A0A1WYM5 P17336 A0A1W4UPD3 B4J1T4 W8C3S7 A0A0K8VAH0 T1E775 J9HGP0 A0A1L7LQD2 A0A182FDJ3 A0A023EUF0 Q16J86 A0A2M4BKY1 A0A1L8E475 A0A182GFN9 G9BQS7 A0A087ZC72 A0A2M4A193 A0A084WKX1 A0A1L8E4H4 K4N1L2 B4LFX2 A0A182IJJ1 E0YL15 A0A2M3YYC6 A0A2M3ZFP9 A0A2M4A1E0 A0A2M4CPT1 A0A2J7REJ5 A0A2M3YYJ7 A0A182WS20 A3E807 A0A182HQG6 A0A182UXY3 A0A1J1I836 A0A2M4CV05 A3E7Z8 A0A2M4CTV7 A0A182LDW5 A0A182PEB9 A3E800 A0A182QZS4 E2AJX6 A0A1B3PEJ4 A0A2M4A121 W5JER1 A0A067RLP1 A0A2M4A0F7 A0A1Q3FPN2 A0A131XJ28 G9C5E5 A0A224YPR9 A0A182N228 A0A224YJA5 A0A224YJ98 L7M7X5 A0A2U8T6B2
PDB
6JNU
E-value=0,
Score=1803
Ontologies
PATHWAY
00380
Tryptophan metabolism - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
04068 FoxO signaling pathway - Bombyx mori (domestic silkworm)
04146 Peroxisome - Bombyx mori (domestic silkworm)
04213 Longevity regulating pathway - multiple species - Bombyx mori (domestic silkworm)
00630 Glyoxylate and dicarboxylate metabolism - Bombyx mori (domestic silkworm)
01100 Metabolic pathways - Bombyx mori (domestic silkworm)
01200 Carbon metabolism - Bombyx mori (domestic silkworm)
04068 FoxO signaling pathway - Bombyx mori (domestic silkworm)
04146 Peroxisome - Bombyx mori (domestic silkworm)
04213 Longevity regulating pathway - multiple species - Bombyx mori (domestic silkworm)
GO
PANTHER
Topology
Subcellular location
Peroxisome
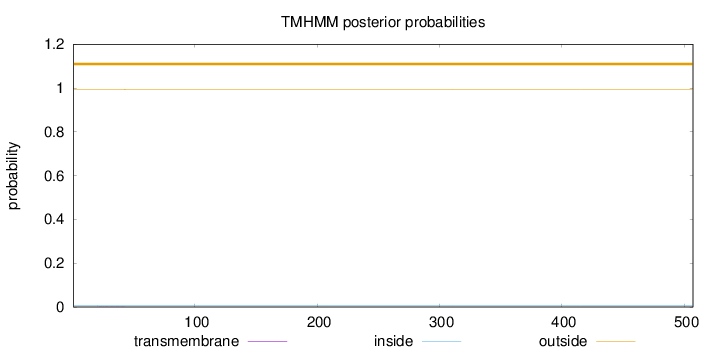
Length:
507
Number of predicted TMHs:
0
Exp number of AAs in TMHs:
0.0247499999999999
Exp number, first 60 AAs:
0.02249
Total prob of N-in:
0.00654
outside
1 - 507
Population Genetic Test Statistics
Pi
1.380674
Theta
2.20691
Tajima's D
-0.997825
CLR
0.007998
CSRT
0.134243287835608
Interpretation
Possibly Positive selection
