Gene
KWMTBOMO00338
Pre Gene Modal
BGIBMGA000527
Annotation
PREDICTED:_facilitated_trehalose_transporter_Tret1-like_isoform_X2_[Bombyx_mori]
Location in the cell
PlasmaMembrane Reliability : 4.973
Sequence
CDS
ATGGCGCTCGACACAGAAGTAGCTGGAGATGGTGCAACGTATGCACATTATGTATACAAAAAGGATTTTCGCACAGCTCTTCCCCAATTTTTAGCTGTGAGCGTAAAGAACCTTCTATTACTGGGCTATGGCATGACCTTGGGTTTTACGACTATACTTATTCCCGCTGTCGAGAAACCAAAAGAGGGCGAAGTTTTATATTTAGAAAAATCTGAGATTTCTTGGATAAGTTCAATAAACTTGATAGTCGTGCCACTGGGGTGTGCAGTATCCGGCATAGTCACGACACCCATTGGTCGCCGACGTTCTATGCAAGTTGTGAACCTTCCATTCTTTATAGCTTGGTTACTATTTCACTTCTCAACCAGTACCGGACATTTATATGCAGCTCTGTTTTTAACGGGTTTAGCTGGAGGTCTCCTTGAAGCACCTGTACTAACGTATGTGGCAGAAATAACACAACCACATCTTCGAGGTGCCTTGACAGCTACTAGTTCTATGTGCATACTAATTGGAGTTTTCACACAGTTTCTGTTTGGTCTCTTTATGTACTGGAGGACAGTGGCACTCGTTAACATTATATTCACAATCTTGGCCGTTATAGCATTATGTTTTGTTCCTGAATCTCCACATTGGCTGGTTGCTAAGAAACGTTACGAAGATGCTAGAAAGAGCTTGCAATGGCTTCGTGGATGGACAGGTCCACATGTCGTGGAACAAGAGTTGCAGGATATCAAAGCTCTATTCAAAAGAGGAAAAGATGTTGATAATACGGAAGAAACGTTGTCAGAAAAGGTATATCAGTATTGTAGAAGAAGTTTCTTACTGCCATTTTTTCTCGTCAGTTTTTCCTTCTTTGTTGGCCATTTCAGCGGCATGACAACTTTACAGACATATGCAGTATCCATATTTCAGATGCTCGAAGCGCCTATTGATAAATACTACGCCACACTCATCCTCGGAATTCTGCAAATACTTGGCTCCGGTGCCTGTGTGCTATTAGTGCACTACACCGGAAAGAGACCTCTCGCATTCTTCTCCACGGGCGTCGCTGGTATATGCTGTATCTTAGTGGCCATCTACGACTTATACGCTCGCACTCACATTATGAGCGGAGTATCTTTGAACGCAGAAAGAGTTGTTACTGATGTCAATGCCACTCTTACCAGTGAAACTGAAGTCTTCGTGAAAAATCCTTATTCATGGATGCCAACAACATTTCTCATGGTACTGGCATTGGCCACGCACACAGGCATTCGGTTGCTCCCTTGGATCTTAATTGGAGAGGTATTTAATGCCAAAACGAGATCCGGTGGTGCCGGATTAGCGAGTGCTGTAGGATATATATTTGGTTTTCTAACTAATAAAATGTACATTTCGATGGTGGATACCCTATCCATTTGGGGAACGTATGGATTTTACGGTATTGTATCCTTAATGGGATGCACGGTGCTATATTTTATTTTACCAGAAACAGAGGGAAAGAAGTTAAATGAAATTGAAAACCATTTCACCGGAATTAGAAAGTTGACAAATCAAGTATACCGGTCGAAGAGAAGACCACAAAACGAGGTGTCAAAAATGCAAGAAATGAAAGGGGCAACAAATCCAACATTCGAAAACGACACTCTGAATATCTAA
Protein
MALDTEVAGDGATYAHYVYKKDFRTALPQFLAVSVKNLLLLGYGMTLGFTTILIPAVEKPKEGEVLYLEKSEISWISSINLIVVPLGCAVSGIVTTPIGRRRSMQVVNLPFFIAWLLFHFSTSTGHLYAALFLTGLAGGLLEAPVLTYVAEITQPHLRGALTATSSMCILIGVFTQFLFGLFMYWRTVALVNIIFTILAVIALCFVPESPHWLVAKKRYEDARKSLQWLRGWTGPHVVEQELQDIKALFKRGKDVDNTEETLSEKVYQYCRRSFLLPFFLVSFSFFVGHFSGMTTLQTYAVSIFQMLEAPIDKYYATLILGILQILGSGACVLLVHYTGKRPLAFFSTGVAGICCILVAIYDLYARTHIMSGVSLNAERVVTDVNATLTSETEVFVKNPYSWMPTTFLMVLALATHTGIRLLPWILIGEVFNAKTRSGGAGLASAVGYIFGFLTNKMYISMVDTLSIWGTYGFYGIVSLMGCTVLYFILPETEGKKLNEIENHFTGIRKLTNQVYRSKRRPQNEVSKMQEMKGATNPTFENDTLNI
Summary
Uniprot
A0A2H1V735
A0A212F446
A0A2A4J6T6
A0A194QVD1
A0A067R6P9
A0A1B6E3R0
+ More
A0A2J7RJ72 A0A1W4XIV3 D6WL92 D4AHX8 A0A1B6KK57 A0A3L8D4G2 A0A154PAA0 A0A1B6G9J5 E2AJT4 A0A1Y1LG83 A0A1B6HRQ7 A0A088ASY5 A0A2A3E5N0 A0A158NN58 E2BGB2 A0A1B6IYD1 A0A310SHC1 A0A1B0GIU9 A0A1Y1KY42 A0A1Y1L2Y1 A0A195C458 A0A0L7RAX7 E0VXC5 A0A232EY14 A0A0C9PJ36 K7J939 A0A195FB02 A0A2H8TUX9 E9IDV1 A0A2S2NJ05 U4U207 J9JXT8 A0A1J1IV84 A0A2S2QNS5 N6U6K2 A0A0N0BFQ3 A0A1Y1MTD9 F4X7I0 A0A1Y1LD79 A0A1Y1L863 A0A026W2D8 A0A1Y1NDF5 A0A151JQ60 A0A1B0D994 A0A195BVJ4 A0A0K8VLW6 A0A034VP51 A0A1Y1K0N9 A0A1Y1JVU2 A0A1I8P3K2 A0A1I8P3J1 A0A1I8P3E8 A0A1Y1NCS6 B0WLM4 A0A0A1X336 A0A0A1XA82 A0A1I8M8T8 A0A1I8M8U1 W8AKQ4 W8AE23 A0A1Q3FRP9 A0A0B4KGW3 Q9VHI9 A0A1W4W2E7 A0A1W4W3V6 B4HKY1 B4JFR1 B4NA62 Q175W6 A0A2M4AGT5 A0A0R1E4T8 B4PTY5 A0A182FS13 A0A2M4AH60 B4M462 A0A0Q9WDV5 B3P1V2 A0A1Y1LY30 A0A182VVZ1 A0A182GGJ1 A0A2M4BJE1 A0A2M4BIQ4
A0A2J7RJ72 A0A1W4XIV3 D6WL92 D4AHX8 A0A1B6KK57 A0A3L8D4G2 A0A154PAA0 A0A1B6G9J5 E2AJT4 A0A1Y1LG83 A0A1B6HRQ7 A0A088ASY5 A0A2A3E5N0 A0A158NN58 E2BGB2 A0A1B6IYD1 A0A310SHC1 A0A1B0GIU9 A0A1Y1KY42 A0A1Y1L2Y1 A0A195C458 A0A0L7RAX7 E0VXC5 A0A232EY14 A0A0C9PJ36 K7J939 A0A195FB02 A0A2H8TUX9 E9IDV1 A0A2S2NJ05 U4U207 J9JXT8 A0A1J1IV84 A0A2S2QNS5 N6U6K2 A0A0N0BFQ3 A0A1Y1MTD9 F4X7I0 A0A1Y1LD79 A0A1Y1L863 A0A026W2D8 A0A1Y1NDF5 A0A151JQ60 A0A1B0D994 A0A195BVJ4 A0A0K8VLW6 A0A034VP51 A0A1Y1K0N9 A0A1Y1JVU2 A0A1I8P3K2 A0A1I8P3J1 A0A1I8P3E8 A0A1Y1NCS6 B0WLM4 A0A0A1X336 A0A0A1XA82 A0A1I8M8T8 A0A1I8M8U1 W8AKQ4 W8AE23 A0A1Q3FRP9 A0A0B4KGW3 Q9VHI9 A0A1W4W2E7 A0A1W4W3V6 B4HKY1 B4JFR1 B4NA62 Q175W6 A0A2M4AGT5 A0A0R1E4T8 B4PTY5 A0A182FS13 A0A2M4AH60 B4M462 A0A0Q9WDV5 B3P1V2 A0A1Y1LY30 A0A182VVZ1 A0A182GGJ1 A0A2M4BJE1 A0A2M4BIQ4
Pubmed
22118469
26354079
24845553
18362917
19820115
20705135
+ More
30249741 20798317 28004739 21347285 20566863 28648823 20075255 21282665 23537049 21719571 24508170 25348373 25830018 25315136 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17994087 17510324 17550304 18057021 26483478
30249741 20798317 28004739 21347285 20566863 28648823 20075255 21282665 23537049 21719571 24508170 25348373 25830018 25315136 24495485 10731132 12537568 12537572 12537573 12537574 16110336 17569856 17569867 17994087 17510324 17550304 18057021 26483478
EMBL
ODYU01000770
SOQ36064.1
AGBW02010446
OWR48501.1
NWSH01002995
PCG67133.1
+ More
KQ461108 KPJ09279.1 KK852665 KDR18984.1 GEDC01004732 JAS32566.1 NEVH01002996 PNF40880.1 KQ971343 EFA03485.2 AB550006 BAI83427.1 GEBQ01028146 JAT11831.1 QOIP01000013 RLU15290.1 KQ434857 KZC08782.1 GECZ01010667 JAS59102.1 GL440100 EFN66250.1 GEZM01062131 JAV70027.1 GECU01030371 GECU01023238 JAS77335.1 JAS84468.1 KZ288361 PBC27005.1 ADTU01020973 ADTU01020974 ADTU01020975 GL448138 EFN85275.1 GECU01015856 JAS91850.1 KQ764421 OAD54523.1 AJWK01017665 GEZM01070708 JAV66323.1 GEZM01070706 JAV66325.1 KQ978344 KYM94958.1 KQ414618 KOC67891.1 DS235830 EEB18031.1 NNAY01001727 OXU23108.1 GBYB01000978 JAG70745.1 AAZX01007103 KQ981693 KYN37561.1 GFXV01005567 MBW17372.1 GL762535 EFZ21271.1 GGMR01004554 MBY17173.1 KB631904 ERL87107.1 ABLF02033085 ABLF02033091 CVRI01000061 CRL04016.1 GGMS01010195 MBY79398.1 APGK01040764 APGK01040765 APGK01040766 APGK01040767 KB740984 ENN76281.1 KQ435798 KOX73422.1 GEZM01023379 JAV88418.1 GL888840 EGI57618.1 GEZM01063752 JAV68977.1 GEZM01063750 GEZM01063749 JAV68978.1 KK107467 EZA50255.1 GEZM01005778 JAV95923.1 KQ978705 KYN29131.1 AJVK01004280 KQ976406 KYM91675.1 GDHF01012433 JAI39881.1 GAKP01015055 JAC43897.1 GEZM01101774 JAV52307.1 GEZM01101773 JAV52308.1 GEZM01006599 JAV95693.1 DS231988 EDS30566.1 GBXI01008563 GBXI01001366 JAD05729.1 JAD12926.1 GBXI01006709 JAD07583.1 GAMC01020136 GAMC01020133 GAMC01020132 JAB86422.1 GAMC01020135 JAB86420.1 GFDL01004766 JAV30279.1 AE014297 AGB95777.1 AAF54318.2 CH480815 EDW42945.1 CH916369 EDV93542.1 CH964232 EDW80705.2 CH477393 EAT41933.1 GGFK01006601 MBW39922.1 CM000160 KRK03219.1 EDW96596.1 KRK03218.1 KRK03222.1 GGFK01006805 MBW40126.1 CH940652 EDW59423.1 KRF78849.1 KRF78850.1 KRF78851.1 KRF78853.1 CH954181 EDV49840.1 GEZM01046374 JAV77280.1 JXUM01061834 JXUM01061835 JXUM01061836 KQ562171 KXJ76503.1 GGFJ01003767 MBW52908.1 GGFJ01003743 MBW52884.1
KQ461108 KPJ09279.1 KK852665 KDR18984.1 GEDC01004732 JAS32566.1 NEVH01002996 PNF40880.1 KQ971343 EFA03485.2 AB550006 BAI83427.1 GEBQ01028146 JAT11831.1 QOIP01000013 RLU15290.1 KQ434857 KZC08782.1 GECZ01010667 JAS59102.1 GL440100 EFN66250.1 GEZM01062131 JAV70027.1 GECU01030371 GECU01023238 JAS77335.1 JAS84468.1 KZ288361 PBC27005.1 ADTU01020973 ADTU01020974 ADTU01020975 GL448138 EFN85275.1 GECU01015856 JAS91850.1 KQ764421 OAD54523.1 AJWK01017665 GEZM01070708 JAV66323.1 GEZM01070706 JAV66325.1 KQ978344 KYM94958.1 KQ414618 KOC67891.1 DS235830 EEB18031.1 NNAY01001727 OXU23108.1 GBYB01000978 JAG70745.1 AAZX01007103 KQ981693 KYN37561.1 GFXV01005567 MBW17372.1 GL762535 EFZ21271.1 GGMR01004554 MBY17173.1 KB631904 ERL87107.1 ABLF02033085 ABLF02033091 CVRI01000061 CRL04016.1 GGMS01010195 MBY79398.1 APGK01040764 APGK01040765 APGK01040766 APGK01040767 KB740984 ENN76281.1 KQ435798 KOX73422.1 GEZM01023379 JAV88418.1 GL888840 EGI57618.1 GEZM01063752 JAV68977.1 GEZM01063750 GEZM01063749 JAV68978.1 KK107467 EZA50255.1 GEZM01005778 JAV95923.1 KQ978705 KYN29131.1 AJVK01004280 KQ976406 KYM91675.1 GDHF01012433 JAI39881.1 GAKP01015055 JAC43897.1 GEZM01101774 JAV52307.1 GEZM01101773 JAV52308.1 GEZM01006599 JAV95693.1 DS231988 EDS30566.1 GBXI01008563 GBXI01001366 JAD05729.1 JAD12926.1 GBXI01006709 JAD07583.1 GAMC01020136 GAMC01020133 GAMC01020132 JAB86422.1 GAMC01020135 JAB86420.1 GFDL01004766 JAV30279.1 AE014297 AGB95777.1 AAF54318.2 CH480815 EDW42945.1 CH916369 EDV93542.1 CH964232 EDW80705.2 CH477393 EAT41933.1 GGFK01006601 MBW39922.1 CM000160 KRK03219.1 EDW96596.1 KRK03218.1 KRK03222.1 GGFK01006805 MBW40126.1 CH940652 EDW59423.1 KRF78849.1 KRF78850.1 KRF78851.1 KRF78853.1 CH954181 EDV49840.1 GEZM01046374 JAV77280.1 JXUM01061834 JXUM01061835 JXUM01061836 KQ562171 KXJ76503.1 GGFJ01003767 MBW52908.1 GGFJ01003743 MBW52884.1
Proteomes
UP000007151
UP000218220
UP000053240
UP000027135
UP000235965
UP000192223
+ More
UP000007266 UP000279307 UP000076502 UP000000311 UP000005203 UP000242457 UP000005205 UP000008237 UP000092461 UP000078542 UP000053825 UP000009046 UP000215335 UP000002358 UP000078541 UP000030742 UP000007819 UP000183832 UP000019118 UP000053105 UP000007755 UP000053097 UP000078492 UP000092462 UP000078540 UP000095300 UP000002320 UP000095301 UP000000803 UP000192221 UP000001292 UP000001070 UP000007798 UP000008820 UP000002282 UP000069272 UP000008792 UP000008711 UP000075920 UP000069940 UP000249989
UP000007266 UP000279307 UP000076502 UP000000311 UP000005203 UP000242457 UP000005205 UP000008237 UP000092461 UP000078542 UP000053825 UP000009046 UP000215335 UP000002358 UP000078541 UP000030742 UP000007819 UP000183832 UP000019118 UP000053105 UP000007755 UP000053097 UP000078492 UP000092462 UP000078540 UP000095300 UP000002320 UP000095301 UP000000803 UP000192221 UP000001292 UP000001070 UP000007798 UP000008820 UP000002282 UP000069272 UP000008792 UP000008711 UP000075920 UP000069940 UP000249989
PRIDE
Pfam
PF00083 Sugar_tr
Interpro
SUPFAM
SSF103473
SSF103473
CDD
ProteinModelPortal
A0A2H1V735
A0A212F446
A0A2A4J6T6
A0A194QVD1
A0A067R6P9
A0A1B6E3R0
+ More
A0A2J7RJ72 A0A1W4XIV3 D6WL92 D4AHX8 A0A1B6KK57 A0A3L8D4G2 A0A154PAA0 A0A1B6G9J5 E2AJT4 A0A1Y1LG83 A0A1B6HRQ7 A0A088ASY5 A0A2A3E5N0 A0A158NN58 E2BGB2 A0A1B6IYD1 A0A310SHC1 A0A1B0GIU9 A0A1Y1KY42 A0A1Y1L2Y1 A0A195C458 A0A0L7RAX7 E0VXC5 A0A232EY14 A0A0C9PJ36 K7J939 A0A195FB02 A0A2H8TUX9 E9IDV1 A0A2S2NJ05 U4U207 J9JXT8 A0A1J1IV84 A0A2S2QNS5 N6U6K2 A0A0N0BFQ3 A0A1Y1MTD9 F4X7I0 A0A1Y1LD79 A0A1Y1L863 A0A026W2D8 A0A1Y1NDF5 A0A151JQ60 A0A1B0D994 A0A195BVJ4 A0A0K8VLW6 A0A034VP51 A0A1Y1K0N9 A0A1Y1JVU2 A0A1I8P3K2 A0A1I8P3J1 A0A1I8P3E8 A0A1Y1NCS6 B0WLM4 A0A0A1X336 A0A0A1XA82 A0A1I8M8T8 A0A1I8M8U1 W8AKQ4 W8AE23 A0A1Q3FRP9 A0A0B4KGW3 Q9VHI9 A0A1W4W2E7 A0A1W4W3V6 B4HKY1 B4JFR1 B4NA62 Q175W6 A0A2M4AGT5 A0A0R1E4T8 B4PTY5 A0A182FS13 A0A2M4AH60 B4M462 A0A0Q9WDV5 B3P1V2 A0A1Y1LY30 A0A182VVZ1 A0A182GGJ1 A0A2M4BJE1 A0A2M4BIQ4
A0A2J7RJ72 A0A1W4XIV3 D6WL92 D4AHX8 A0A1B6KK57 A0A3L8D4G2 A0A154PAA0 A0A1B6G9J5 E2AJT4 A0A1Y1LG83 A0A1B6HRQ7 A0A088ASY5 A0A2A3E5N0 A0A158NN58 E2BGB2 A0A1B6IYD1 A0A310SHC1 A0A1B0GIU9 A0A1Y1KY42 A0A1Y1L2Y1 A0A195C458 A0A0L7RAX7 E0VXC5 A0A232EY14 A0A0C9PJ36 K7J939 A0A195FB02 A0A2H8TUX9 E9IDV1 A0A2S2NJ05 U4U207 J9JXT8 A0A1J1IV84 A0A2S2QNS5 N6U6K2 A0A0N0BFQ3 A0A1Y1MTD9 F4X7I0 A0A1Y1LD79 A0A1Y1L863 A0A026W2D8 A0A1Y1NDF5 A0A151JQ60 A0A1B0D994 A0A195BVJ4 A0A0K8VLW6 A0A034VP51 A0A1Y1K0N9 A0A1Y1JVU2 A0A1I8P3K2 A0A1I8P3J1 A0A1I8P3E8 A0A1Y1NCS6 B0WLM4 A0A0A1X336 A0A0A1XA82 A0A1I8M8T8 A0A1I8M8U1 W8AKQ4 W8AE23 A0A1Q3FRP9 A0A0B4KGW3 Q9VHI9 A0A1W4W2E7 A0A1W4W3V6 B4HKY1 B4JFR1 B4NA62 Q175W6 A0A2M4AGT5 A0A0R1E4T8 B4PTY5 A0A182FS13 A0A2M4AH60 B4M462 A0A0Q9WDV5 B3P1V2 A0A1Y1LY30 A0A182VVZ1 A0A182GGJ1 A0A2M4BJE1 A0A2M4BIQ4
PDB
4ZWC
E-value=9.14973e-13,
Score=179
Ontologies
GO
Topology
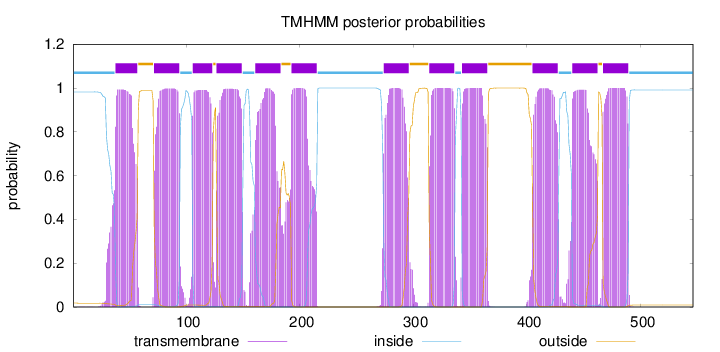
Length:
546
Number of predicted TMHs:
12
Exp number of AAs in TMHs:
262.93056
Exp number, first 60 AAs:
21.24104
Total prob of N-in:
0.98222
POSSIBLE N-term signal
sequence
inside
1 - 37
TMhelix
38 - 57
outside
58 - 71
TMhelix
72 - 94
inside
95 - 105
TMhelix
106 - 123
outside
124 - 126
TMhelix
127 - 149
inside
150 - 160
TMhelix
161 - 183
outside
184 - 192
TMhelix
193 - 215
inside
216 - 273
TMhelix
274 - 296
outside
297 - 313
TMhelix
314 - 336
inside
337 - 342
TMhelix
343 - 365
outside
366 - 404
TMhelix
405 - 427
inside
428 - 439
TMhelix
440 - 462
outside
463 - 466
TMhelix
467 - 489
inside
490 - 546
Population Genetic Test Statistics
Pi
14.867727
Theta
13.276565
Tajima's D
-2.088761
CLR
0.182336
CSRT
0.00844957752112394
Interpretation
Uncertain
